Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa
- PMID: 10611362
- PMCID: PMC24797
- DOI: 10.1073/pnas.96.26.15202
Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa
Abstract
Identification of host factors that interact with pathogens is crucial to an understanding of infectious disease, but direct screening for host mutations to aid in this task is not feasible in mammals. The nematode Caenorhabditis elegans is a genetically tractable alternative for investigating the pathogenic bacterium Pseudomonas aeruginosa. A P. aeruginosa toxin, produced at high cell density under control of the quorum-sensing regulators LasR and RhlR, rapidly and lethally paralyzes C. elegans. Loss-of-function mutations in C. elegans egl-9, a gene required for normal egg laying, confer strong resistance to the paralysis. Thus, activation of EGL-9 or of a pathway that includes it may lead to the paralysis. The molecular identity of egl-9 was determined by transformation rescue and DNA sequencing. A mammalian homologue of EGL-9 is expressed in tissues in which exposure to P. aeruginosa could have clinical effects.
Figures
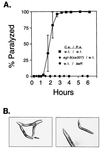
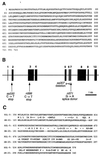
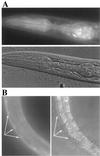
Comment in
-
Novel antimicrobial targets from combined pathogen and host genetics.Proc Natl Acad Sci U S A. 2000 Feb 1;97(3):958-9. doi: 10.1073/pnas.97.3.958. Proc Natl Acad Sci U S A. 2000. PMID: 10655466 Free PMC article. No abstract available.
References
-
- Caenorhabditis elegans Sequencing Consortium. Science. 1998;282:2012–2018. - PubMed
-
- Campra M, Bendinelli M, Friedman H, editors. Pseudomonas aeruginosa as an Opportunistic Pathogen. New York: Plenum; 1993.
-
- Pollack M. In: Infectious Diseases. 2nd Ed. Gorbach S L, Bartlett J G, Blacklow N R, editors. Philadelphia: Saunders; 1998. pp. 1824–1837.
-
- Fuqua C, Winans S C, Greenberg E P. Annu Rev Microbiol. 1996;50:727–751. - PubMed
-
- Mahajan-Miklos S, Tan M-W, Rahme L G, Ausubel F M. Cell. 1999;96:47–56. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

