A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription
- PMID: 10611387
- PMCID: PMC24822
- DOI: 10.1073/pnas.96.26.15348
A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription
Abstract
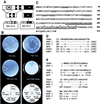
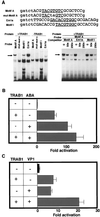
The transcription factor VP1 regulates maturation and dormancy in plant seeds by activating genes responsive to the stress hormone abscisic acid (ABA). Although activation involves ABA-responsive elements (ABREs), VP1 itself does not specifically bind ABREs. Instead, we have identified and cloned a basic region leucine zipper (bZIP) factor, TRAB1, that interacts with both VP1 and ABREs. Transcription from a chimeric promoter with GAL4-binding sites was ABA-inducible if cells expressed a GAL4 DNA-binding domain::TRAB1 fusion protein. Results indicate that TRAB1 is a true trans-acting factor involved in ABA-regulated transcription and reveal a molecular mechanism for the VP1-dependent, ABA-inducible transcription that controls maturation and dormancy in plant embryos.
Figures
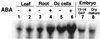

References
-
- Giraudat J, Parcy F, Bertauche N, Gosti F, Leung J, Morris P C, Bouvier-Durand M, Vartanian N. Plant Mol Biol. 1994;26:1557–1577. - PubMed
-
- McCarty D R. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:71–93.
-
- Busk P K, Pages M. Plant Mol Biol. 1998;37:425–435. - PubMed
-
- Hattori T, Terada T, Hamasuna S. Plant J. 1995;7:913–925. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

