Assessment of polymorphism in zebrafish mapping strains
- PMID: 10613846
- PMCID: PMC311009
- DOI: 10.1101/gr.9.12.1231
Assessment of polymorphism in zebrafish mapping strains
Abstract
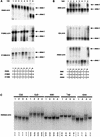
To assess the level of heterozygosity within two commonly used inbred mapping zebrafish strains, C32 and SJD, we genotyped polymorphic CA-repeat markers randomly dispersed throughout the zebrafish genome. (For clarity purposes we will primarily use the term polymorphic to define polymorphism between strains, and the term heterozygous to address heterogeneity within a strain.) Eight male individuals each from C32 and SJD stocks were typed for 235 and 183 markers, respectively. Over 90% of the markers typed were polymorphic between these two strains. We found a limited number of heterozygous markers persisting in clusters within each inbred line. In the SJD strain, these were mainly limited to a few telomeric regions or regions otherwise distant from centromeres. As expected, centromeric regions were homozygous in the SJD strain, consistent with its derivation from a single half-tetrad individual. In contrast, heterozygous clusters were distributed randomly throughout the genome in the C32 strain, and these clusters could be detected with linked polymorphic markers. Nevertheless, most regions of the C32 strain are homozygous for CA-repeat markers in current stocks. This identification of the heterozygous regions within C32 and SJD lines should permit rapid fixation of these remaining regions in future generations of inbreeding. In addition, we established levels of polymorphism between the inbred, C32 and SJD, strains and three other commonly used strains, the *AB, WIK, and Florida wild type (hereafter referred as EKK), with CA-repeat markers as well as SSCP polymorphisms. These data will maximize the use of these strains in mapping experiments.
Figures
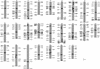
References
-
- Gates MA, Kim L, Egan ES, Cardozo T, Sirotkin HI, Dougan ST, Lashkari D, Abagyan R, Schier AF, Talbot WS. A genetic linkage map for zebrafish: Comparative analysis and localization of genes and expressed sequences. Genome Res. 1999;9:348–359. - PubMed
-
- Goff DJ, Galvin K, Katz H, Westerfield M, Lander ES, Tabin CJ. Identification of polymorphic simple sequence repeats in the genome of the zebrafish. Genomics. 1992;14:200–202. - PubMed
-
- Johnson SL, Zon LI. Genetic backgrounds and some standard stocks and strains used in zebrafish developmental biology and genetics. Methods Cell Biol. 1999;60:357–359. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
