Alternative splicing regulates the subcellular localization of A-kinase anchoring protein 18 isoforms
- PMID: 10613906
- PMCID: PMC2174236
- DOI: 10.1083/jcb.147.7.1481
Alternative splicing regulates the subcellular localization of A-kinase anchoring protein 18 isoforms
Abstract
The cAMP-dependent protein kinase (PKA) is localized to specific subcellular compartments by association with A-kinase anchoring proteins (AKAPs). AKAPs are a family of functionally related proteins that bind the regulatory (R) subunit of PKA with high affinity and target the kinase to specific subcellular organelles. Recently, AKAP18, a low molecular weight plasma membrane AKAP that facilitates PKA-mediated phosphorylation of the L-type Ca(2+) channel, was cloned. We now report the cloning of two additional isoforms of AKAP18, which we have designated AKAP18beta and AKAP18gamma, that arise from alternative mRNA splicing. The AKAP18 isoforms share a common R subunit binding site, but have distinct targeting domains. The original AKAP18 (renamed AKAP18alpha) and AKAP18beta target the plasma membrane when expressed in HEK-293 cells, while AKAP18gamma is cytosolic. When expressed in epithelial cells, AKAP18alpha is targeted to lateral membranes, whereas AKAP18beta is accumulated at the apical membrane. A 23-amino acid insert, following the plasma membrane targeting domain, facilitates the association of AKAP18beta with the apical membrane. The data suggest that AKAP18 isoforms are differentially targeted to modulate distinct intracellular signaling events. Furthermore, the data suggest that plasma membrane AKAPs may be targeted to subdomains of the cell surface, adding additional specificity in intracellular signaling.
Figures




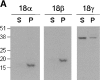
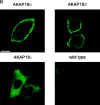
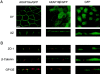
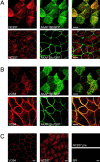

References
-
- Angelo R., Rubin C.S. Molecular characterization of an anchor protein (AKAPCE) that binds the RI subunit (RCE) of type I protein kinase A from Caenorhabditis elegans . J. Biol. Chem. 1998;273:14633–14643. - PubMed
-
- Brown D.A., London E. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 1998;14:111–136. - PubMed
-
- Burton K.A., Johnson B.D., Hausken Z.E., Westenbroek R.E., Idzerda R.L., Scheuer T., Scott J.D., Catterall W.A., McKnight G.S. Type II regulatory subunits are not required for the anchoring-dependent modulation of Ca2+ channel activity by cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 1997;94:11067–11072. - PMC - PubMed
-
- Carr D.W., Stofko-Hahn R.E., Fraser I.D., Bishop S.M., Acott T.S., Brennan R.G., Scott J.D. Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J. Biol. Chem. 1991;266:14188–14192. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

