Typing of Candida glabrata in clinical isolates by comparative sequence analysis of the cytochrome c oxidase subunit 2 gene distinguishes two clusters of strains associated with geographical sequence polymorphisms
- PMID: 10618092
- PMCID: PMC88700
- DOI: 10.1128/JCM.38.1.227-235.2000
Typing of Candida glabrata in clinical isolates by comparative sequence analysis of the cytochrome c oxidase subunit 2 gene distinguishes two clusters of strains associated with geographical sequence polymorphisms
Abstract
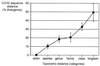
We tested whether comparative sequence analysis of the mitochondrion-encoded cytochrome c oxidase subunit 2 gene (COX2) could be used to distinguish intraspecific variants of Candida glabrata. Mitochondrial genes are suitable for investigation of close phylogenetic relationships because they evolve much faster than nuclear genes, which in general exhibit very limited intraspecific variation. For this survey we used 11 clinical isolates of C. glabrata from three different geographical locations in Brazil, 10 isolates from one location in the United States, 1 American Type Culture Collection strain as an internal control, and the published sequence of strain CBS 138. The complete coding region of COX2 was amplified from total cellular DNA, and both strands were sequenced twice for each strain. These sequences were aligned with published sequences from other fungi, and the numbers of substitutions and phylogenetic relationships were determined. Typing of these strains was done by using 17 substitutions, with 8 being nonsynonymous and 9 being synonymous. Also, cDNAs made from purified mitochondrial polyadenylated RNA were sequenced to confirm that our sequences correspond to the expressed copies and not nuclear pseudogenes and that a frameshift mutation exists in the 3' end of the coding region (position 673) relative to the Saccharomyces cerevisiae sequence and the previously published C. glabrata sequence. We estimated the average evolutionary rate of COX2 to be 11.4% sequence divergence/10(8) years and that phylogenetic relationships of yeasts based on these sequences are consistent with rRNA sequence data. Our analysis of COX2 sequences enables typing of C. glabrata strains based on 13 haplotypes and suggests that positions 51 and 519 indicate a geographical polymorphism that discriminates strains isolated in the United States and strains isolated in Brazil. This provides for the first time a means of typing of Candida strains that cause infections by use of direct sequence comparisons and the associated divergence estimates.
Figures
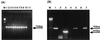





References
-
- Brown W M, Prager E M, Wang A, Wilson A C. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol. 1982;18:225–239. - PubMed
-
- Clark-Walker G D. Contrasting mutation rates in mitochondrial and nuclear genes of yeasts versus mammals. Curr Genet. 1991;20:195–198. - PubMed
-
- Clark-Walker G D, Weiller G F. The structure of the small mitochondrial DNA of Kuyveromyces thermotolerans is likely to reflect ancestral gene order in fungi. J Mol Evol. 1994;38:593–601. - PubMed
-
- Collura R V, Auerbach M R, Stewart C-B. A quick, direct method that can differentiate expressed mitochondrial genes from their nuclear pseudogenes. Curr Biol. 1996;6:1337–1339. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

