Genetic diversity of Vibrio cholerae in Chesapeake Bay determined by amplified fragment length polymorphism fingerprinting
- PMID: 10618215
- PMCID: PMC91797
- DOI: 10.1128/AEM.66.1.140-147.2000
Genetic diversity of Vibrio cholerae in Chesapeake Bay determined by amplified fragment length polymorphism fingerprinting
Abstract
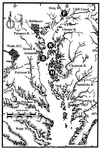
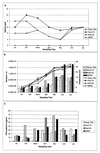
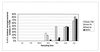
Vibrio cholerae is indigenous to the aquatic environment, and serotype non-O1 strains are readily isolated from coastal waters. However, in comparison with intensive studies of the O1 group, relatively little effort has been made to analyze the population structure and molecular evolution of non-O1 V. cholerae. In this study, high-resolution genomic DNA fingerprinting, amplified fragment length polymorphism (AFLP), was used to characterize the temporal and spatial genetic diversity of 67 V. cholerae strains isolated from Chesapeake Bay during April through July 1998, at four different sampling sites. Isolation of V. cholerae during the winter months (January through March) was unsuccessful, as observed in earlier studies (J. H. L. Kaper, R. R. Colwell, and S. W. Joseph, Appl. Environ. Microbiol. 37:91-103, 1979). AFLP fingerprints subjected to similarity analysis yielded a grouping of isolates into three large clusters, reflecting time of the year when the strains were isolated. April and May isolates were closely related, while July isolates were genetically diverse and did not cluster with the isolates obtained earlier in the year. The results suggest that the population structure of V. cholerae undergoes a shift in genotype that is linked to changes in environmental conditions. From January to July, the water temperature increased from 3 degrees C to 27.5 degrees C, bacterial direct counts increased nearly an order of magnitude, and the chlorophyll a concentration tripled (or even quadrupled at some sites). No correlation was observed between genetic similarity among isolates and geographical source of isolation, since isolates found at a single sampling site were genetically diverse and genetically identical isolates were found at several of the sampling sites. Thus, V. cholerae populations may be transported by surface currents throughout the entire Bay, or, more likely, similar environmental conditions may be selected for a specific genotype. The dynamic nature of the population structure of this bacterial species in Chesapeake Bay provides new insight into the ecology and molecular evolution of V. cholerae in the natural environment.
Figures


Similar articles
-
Genetic diversity of clinical and environmental isolates of Vibrio cholerae determined by amplified fragment length polymorphism fingerprinting.Appl Environ Microbiol. 2000 Jan;66(1):148-53. doi: 10.1128/AEM.66.1.148-153.2000. Appl Environ Microbiol. 2000. PMID: 10618216 Free PMC article.
-
Environmental and epidemiological surveillance of Vibrio cholerae in a cholera-endemic region in India with freshwater environs.J Appl Microbiol. 2012 Jan;112(1):225-37. doi: 10.1111/j.1365-2672.2011.05191.x. Epub 2011 Nov 29. J Appl Microbiol. 2012. PMID: 22054549
-
Amplified fragment length polymorphism of clinical and environmental Vibrio cholerae from a freshwater environment in a cholera-endemic area, India.BMC Infect Dis. 2011 Sep 22;11:249. doi: 10.1186/1471-2334-11-249. BMC Infect Dis. 2011. PMID: 21936962 Free PMC article.
-
Predictability of Vibrio cholerae in Chesapeake Bay.Appl Environ Microbiol. 2003 May;69(5):2773-85. doi: 10.1128/AEM.69.5.2773-2785.2003. Appl Environ Microbiol. 2003. PMID: 12732548 Free PMC article.
-
Inland cholera in freshwater environs of north India.Vaccine. 2020 Feb 29;38 Suppl 1:A63-A72. doi: 10.1016/j.vaccine.2019.06.038. Epub 2019 Jul 3. Vaccine. 2020. PMID: 31279566 Review.
Cited by
-
Quantitative microbial risk assessment of pathogenic vibrios in marine recreational waters of southern california.Appl Environ Microbiol. 2013 Jan;79(1):294-302. doi: 10.1128/AEM.02674-12. Epub 2012 Oct 26. Appl Environ Microbiol. 2013. PMID: 23104412 Free PMC article.
-
Detection and transformation of genome segments that differ within a coastal population of Vibrio cholerae strains.Appl Environ Microbiol. 2007 Jun;73(11):3695-704. doi: 10.1128/AEM.02735-06. Epub 2007 Apr 20. Appl Environ Microbiol. 2007. PMID: 17449699 Free PMC article.
-
Non-O1/non-O139 Vibrio cholerae carrying multiple virulence factors and V. cholerae O1 in the Chesapeake Bay, Maryland.Appl Environ Microbiol. 2015 Mar;81(6):1909-18. doi: 10.1128/AEM.03540-14. Epub 2015 Jan 2. Appl Environ Microbiol. 2015. PMID: 25556194 Free PMC article.
-
Serogroup conversion of Vibrio cholerae in aquatic reservoirs.PLoS Pathog. 2007 Jun;3(6):e81. doi: 10.1371/journal.ppat.0030081. PLoS Pathog. 2007. PMID: 17559304 Free PMC article.
-
Pandemic spread of cholera: genetic diversity and relationships within the seventh pandemic clone of Vibrio cholerae determined by amplified fragment length polymorphism.J Clin Microbiol. 2002 Jan;40(1):172-81. doi: 10.1128/JCM.40.1.172-181.2002. J Clin Microbiol. 2002. PMID: 11773113 Free PMC article.
References
-
- Albert M J, Siddique A K, Islam M S, Faruque A S G, Ansaruzzaman M, Faruque S M, Sack R B. A large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet. 1993;341:704. - PubMed
-
- Arias C, Verdonck L, Swings J, Aznar R, Garay E. A polyphasic approach to study the intraspecific diversity amongst Vibrio vulnificus isolates. Syst Appl Microbiol. 1997;20:622–633.
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1995.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

