Development of methods to detect "Norwalk-like viruses" (NLVs) and hepatitis A virus in delicatessen foods: application to a food-borne NLV outbreak
- PMID: 10618226
- PMCID: PMC91808
- DOI: 10.1128/AEM.66.1.213-218.2000
Development of methods to detect "Norwalk-like viruses" (NLVs) and hepatitis A virus in delicatessen foods: application to a food-borne NLV outbreak
Abstract
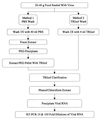
"Norwalk-like viruses" (NLVs) and hepatitis A virus (HAV) are the most common causes of virus-mediated food-borne illness. Epidemiological investigations of outbreaks associated with these viruses have been hindered by the lack of available methods for the detection of NLVs and HAV in foodstuffs. Although reverse transcription (RT)-PCR methods have been useful in detecting NLVs and HAV in bivalve mollusks implicated in outbreaks, to date such methods have not been available for other foods. To address this need, we developed a method to detect NLVs and HAV recovered from food samples. The method involves washing of food samples with a guanidinium-phenol-based reagent, extraction with chloroform, and precipitation in isopropanol. Recovered viral RNA is amplified with HAV- or NLV-specific primers in RT-PCRs, using a viral RNA internal standard control to identify potential sample inhibition. By this method, 10 to 100 PCR units (estimated to be equivalent to 10(2) to 10(3) viral genome copies) of HAV and Norwalk virus seeded onto ham, turkey, and roast beef were detected. The method was applied to food samples implicated in an NLV-associated outbreak at a university cafeteria. Sliced deli ham was positive for a genogroup II NLV as determined by using both polymerase- and capsid-specific primers and probes. Sequence analysis of the PCR-amplified capsid region of the genome indicated that the sequence was identical to the sequence from virus detected in the stools of ill students. The developed method is rapid, simple, and efficient.
Figures


References
-
- Ando T, Jin Q, Gentsch J R, Monroe S S, Noel J S, Dowell S F, Cicirello H G, Kohn M A, Glass R I. Epidemiologic applications of novel molecular methods to detect and differentiate small round structured viruses (Norwalk-like viruses) J Med Virol. 1995;47:145–152. - PubMed
-
- Beller M, Ellis A, Lee S H, Drebot M A, Jenkerson S A, Funk E, Sobsey M D, Simmons O D I, Monroe S S, Ando T, Noel J, Petric M, Middaugh J P, Spika J S. Outbreak of viral gastroenteritis due to a contaminated well. International consequences. JAMA. 1997;278:563–568. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

