In situ hybridization of Prochlorococcus and Synechococcus (marine cyanobacteria) spp. with RRNA-targeted peptide nucleic acid probes
- PMID: 10618237
- PMCID: PMC91819
- DOI: 10.1128/AEM.66.1.284-289.2000
In situ hybridization of Prochlorococcus and Synechococcus (marine cyanobacteria) spp. with RRNA-targeted peptide nucleic acid probes
Abstract
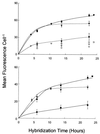
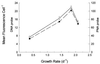
A simple method for whole-cell hybridization using fluorescently labeled rRNA-targeted peptide nucleic acid (PNA) probes was developed for use in marine cyanobacterial picoplankton. In contrast to established protocols, this method is capable of detecting rRNA in Prochlorococcus, the most abundant unicellular marine cyanobacterium. Because the method avoids the use of alcohol fixation, the chlorophyll content of Prochlorococcus cells is preserved, facilitating the identification of these cells in natural samples. PNA probe-conferred fluorescence was measured flow cytometrically and was always significantly higher than that of the negative control probe, with positive/negative ratio varying between 4 and 10, depending on strain and culture growth conditions. Prochlorococcus cells from open ocean samples were detectable with this method. RNase treatment reduced probe-conferred fluorescence to background levels, demonstrating that this signal was in fact related to the presence of rRNA. In another marine cyanobacterium, Synechococcus, in which both PNA and oligonucleotide probes can be used in whole-cell hybridizations, the magnitude of fluorescence from the former was fivefold higher than that from the latter, although the positive/negative ratio was comparable for both probes. In Synechococcus cells growing at a range of growth rates (and thus having different rRNA concentrations per cell), the PNA- and oligonucleotide-derived signals were highly correlated (r = 0.99). The chemical nature of PNA, the sensitivity of PNA-RNA binding to single-base-pair mismatches, and the preservation of cellular integrity by this method suggest that it may be useful for phylogenetic probing of whole cells in the natural environment.
Figures




Similar articles
-
Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides.Microbiology (Reading). 2001 Jul;147(Pt 7):1731-1744. doi: 10.1099/00221287-147-7-1731. Microbiology (Reading). 2001. PMID: 11429451
-
In situ identification of cyanobacteria with horseradish peroxidase-labeled, rRNA-targeted oligonucleotide probes.Appl Environ Microbiol. 1999 Mar;65(3):1259-67. doi: 10.1128/AEM.65.3.1259-1267.1999. Appl Environ Microbiol. 1999. PMID: 10049892 Free PMC article.
-
[Identification of Mycobacterium species from BACTEC MGIT™ positive cultures with Oligo-FISH and PNA-FISH methods].Mikrobiyol Bul. 2014 Jul;48(3):385-401. doi: 10.5578/mb.7876. Mikrobiyol Bul. 2014. PMID: 25052105 Turkish.
-
Phylogenetic identification and in situ detection of individual microbial cells without cultivation.Microbiol Rev. 1995 Mar;59(1):143-69. doi: 10.1128/mr.59.1.143-169.1995. Microbiol Rev. 1995. PMID: 7535888 Free PMC article. Review.
-
The peptide nucleic acids: a new way for chromosomal investigation on isolated cells?Hum Reprod. 2004 Sep;19(9):1946-51. doi: 10.1093/humrep/deh386. Epub 2004 Jun 30. Hum Reprod. 2004. PMID: 15229198 Review.
Cited by
-
Targeting species-specific low-affinity 16S rRNA binding sites by using peptide nucleic acids for detection of Legionellae in biofilms.Appl Environ Microbiol. 2006 Aug;72(8):5453-62. doi: 10.1128/AEM.02918-05. Appl Environ Microbiol. 2006. PMID: 16885298 Free PMC article.
-
Quantitative rRNA-targeted solution-based hybridization assay using peptide nucleic acid molecular beacons.Appl Environ Microbiol. 2008 Dec;74(23):7297-305. doi: 10.1128/AEM.01002-08. Epub 2008 Sep 26. Appl Environ Microbiol. 2008. PMID: 18820054 Free PMC article.
-
Phylogeny and biogeography of cyanobacteria and their produced toxins.Mar Drugs. 2013 Nov 1;11(11):4350-69. doi: 10.3390/md11114350. Mar Drugs. 2013. PMID: 24189276 Free PMC article. Review.
-
Mesozooplankton taurine production and prokaryotic uptake in the northern Adriatic Sea.Limnol Oceanogr. 2020 Nov;65(11):2730-2747. doi: 10.1002/lno.11544. Epub 2020 Jun 25. Limnol Oceanogr. 2020. PMID: 33664530 Free PMC article.
-
Quantification of Marine Picocyanobacteria on Water Column Particles and in Sediments Using Real-Time PCR Reveals Their Role in Carbon Export.mSphere. 2022 Dec 21;7(6):e0049922. doi: 10.1128/msphere.00499-22. Epub 2022 Dec 6. mSphere. 2022. PMID: 36472446 Free PMC article.
References
-
- Bidigare R R, Ondrusek M E. Spatial and temporal variability of phytoplankton pigment distributions in the central equatorial Pacific Ocean. Deep-Sea Res II. 1996;43:809–833.
-
- Binder B J, Chisholm S W, Olson R J, Frankel S L, Worden A Z. Dynamics of pico-phytoplankton, ultra-phytoplankton, and bacteria in the central equatorial Pacific. Deep-Sea Res Part II. 1996;43:907–931.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

