High rate of recombination throughout the human immunodeficiency virus type 1 genome
- PMID: 10627533
- PMCID: PMC111457
- DOI: 10.1128/jvi.74.3.1234-1240.2000
High rate of recombination throughout the human immunodeficiency virus type 1 genome
Abstract
The diploid nature of human immunodeficiency virus type 1 (HIV-1) indicates that recombination serves a central function in virus replication and evolution. Previously, while examining the nature of obligatory primer strand transfers during reverse transcription, a high rate of recombination was observed at the ends of the viral genome within the viral long terminal repeats, prompting the following question: does recombination occur at a high rate throughout the genome? To address this question, two vectors based upon different strains of HIV-1 were utilized. The vectors were comprised predominantly of autologous HIV-1 sequence and were approximately the same size as the parental genome. The proviral progeny of heterodimeric virions were analyzed after a single cycle of replication, and the sequence heterogeneity between the two strains allowed direct examination of recombination crossovers. The results obtained indicate that HIV-1 undergoes approximately two to three recombination events per genome per replication cycle. These results imply that both HIV-1 RNAs are typically utilized during reverse transcription and that recombination is an important aspect of HIV-1 replication.
Figures


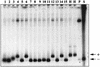
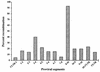
References
-
- Blackard J T, Renjifo B R, Mwakagile D, Montano M A, Fawzi W W, Essex M. Transmission of human immunodeficiency type 1 viruses with intersubtype recombinant long terminal repeat sequences. Virology. 1999;254:220–225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

