An African swine fever virus ERV1-ALR homologue, 9GL, affects virion maturation and viral growth in macrophages and viral virulence in swine
- PMID: 10627538
- PMCID: PMC111462
- DOI: 10.1128/jvi.74.3.1275-1285.2000
An African swine fever virus ERV1-ALR homologue, 9GL, affects virion maturation and viral growth in macrophages and viral virulence in swine
Abstract
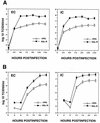
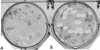
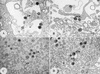
The African swine fever virus (ASFV) genome contains a gene, 9GL, with similarity to yeast ERV1 and ALR genes. ERV1 has been shown to function in oxidative phosphorylation and in cell growth, while ALR has hepatotrophic activity. 9GL encodes a protein of 119 amino acids and was highly conserved at both nucleotide and amino acid levels among all ASFV field isolates examined. Monospecific rabbit polyclonal antibody produced to a glutathione S-transferase-9GL fusion protein specifically immunoprecipitated a 14-kDa protein from macrophage cell cultures infected with the ASFV isolate Malawi Lil-20/1 (MAL). Time course analysis and viral DNA synthesis inhibitor experiments indicated that p14 was a late viral protein. A 9GL gene deletion mutant of MAL (Delta9GL), exhibited a growth defect in macrophages of approximately 2 log(10) units and had a small-plaque phenotype compared to either a revertant (9GL-R) or the parental virus. 9GL affected normal virion maturation; virions containing acentric nucleoid structures comprised 90 to 99% of all virions observed in Delta9GL-infected macrophages. The Delta9GL virus was markedly attenuated in swine. In contrast to 9GL-R infection, where mortality was 100%, all Delta9GL-infected animals survived infection. With the exception of a transient fever response in some animals, Delta9GL-infected animals remained clinically normal and exhibited significant 100- to 10,000-fold reductions in viremia titers. All pigs previously infected with Delta9GL survived infection when subsequently challenged with a lethal dose of virulent parental MAL. Thus, ASFV 9GL gene deletion mutants may prove useful as live-attenuated ASF vaccines.
Figures




References
-
- Afonso C L, Alcaraz C, Brun A, Sussman M D, Onisk D V, Escribano J M, Rock D L. Characterization of p30, a highly antigenic membrane and secreted protein of African swine fever virus. Virology. 1992;189:368–373. - PubMed
-
- Afonso C L, Zsak L, Carrillo C, Borca M V, Rock D L. African swine fever virus NL gene is not required for virus virulence. J Gen Virol. 1998;79:2543–2547. - PubMed
-
- Alcaraz C, Pasamontes B, Ruiz G F, Escribano J M. African swine fever virus-induced proteins on the plasma membranes of infected cells. Virology. 1989;168:406–408. - PubMed
-
- Borca M V, Irusta P M, Kutish G F, Carrillo C, Afonso C L, Burrage T, Neilan J G, Rock D L. A structural DNA binding protein of African swine fever virus with similarity to bacterial histone-like proteins. Arch Virol. 1996;141:301–313. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

