Two patches of amino acids on the E2 DNA binding domain define the surface for interaction with E1
- PMID: 10627562
- PMCID: PMC111486
- DOI: 10.1128/jvi.74.3.1506-1512.2000
Two patches of amino acids on the E2 DNA binding domain define the surface for interaction with E1
Abstract
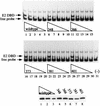
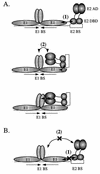
The E1 and E2 proteins from bovine papillomavirus bind cooperatively to the viral origin of DNA replication (ori), forming a complex which is essential for initiation of DNA replication. Cooperative binding has two components, in which (i) the DNA binding domains (DBDs) of the two proteins interact with each other and (ii) the E2 transactivation domain interacts with the helicase domain of E1. By generating specific point mutations in the DBD of E2, we have defined two patches of amino acids that are involved in the interaction with the E1 DBD. These same mutations, when introduced into the viral genome, result in severely reduced replication of the viral genome, as well as failure to transform mouse cells in tissue culture. Thus, the interaction between the E1 and E2 DBDs is important for the establishment of the viral genome as an episome and most likely contributes to the formation of a preinitiation complex on the viral ori.
Figures


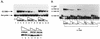


References
-
- Chen L, Glover J N M, Hogan P G, Rao A, Harrison S C. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

