Tight control of respiration by NADH dehydrogenase ND5 subunit gene expression in mouse mitochondria
- PMID: 10629037
- PMCID: PMC85197
- DOI: 10.1128/MCB.20.3.805-815.2000
Tight control of respiration by NADH dehydrogenase ND5 subunit gene expression in mouse mitochondria
Abstract
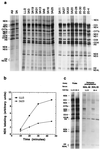
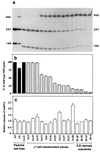
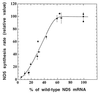
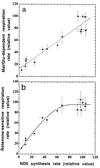
A mouse cell variant carrying in heteroplasmic form a nonsense mutation in the mitochondrial DNA-encoded ND5 subunit of the respiratory NADH dehydrogenase has been isolated and characterized. The derivation from this mutant of a large number of cell lines containing between 4 and 100% of the normal number of wild-type ND5 genes has allowed an analysis of the genetic and functional thresholds operating in mouse mitochondria. In wild-type cells, approximately 40% of the ND5 mRNA level was in excess of that required for ND5 subunit synthesis. However, in heteroplasmic cells, the functional mRNA level decreased in proportion to the number of wild-type ND5 genes over a 25-fold range, pointing to the lack of any compensatory increase in rate of transcription and/or stability of mRNA. Most strikingly, the highest ND5 synthesis rate was just sufficient to support the maximum NADH dehydrogenase-dependent respiration rate, with no upregulation of translation occurring with decreasing wild-type mRNA levels. These results indicate that, despite the large excess of genetic potential of the mammalian mitochondrial genome, respiration is tightly regulated by ND5 gene expression.
Figures



References
-
- Ammini C V, Hauswirth W W. Mitochondrial gene expression is regulated at the level of transcription during early embryogenesis of Xenopus laevis. J Biol Chem. 1999;274:6265–6271. - PubMed
-
- Attardi G. Animal mitochondrial DNA: an extreme example of genetic economy. Int Rev Cytol. 1985;93:93–145. - PubMed
-
- Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. - PubMed
-
- Attardi G, Chomyn A, Loguercio P. Evidence for translational control of mitochondrial gene expression in rat muscle and brain synaptosome mitochondria. In: Benzi G, editor. Advances in myochemistry. Vol. 2. London, England: John Libbey Eurotext, Ltd.; 1989. pp. 55–64.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
