The homeodomain of PDX-1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter
- PMID: 10629047
- PMCID: PMC85207
- DOI: 10.1128/MCB.20.3.900-911.2000
The homeodomain of PDX-1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter
Abstract
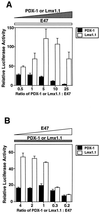
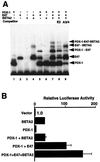
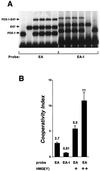
Activation of insulin gene transcription specifically in the pancreatic beta cells depends on multiple nuclear proteins that interact with each other and with sequences on the insulin gene promoter to build a transcriptional activation complex. The homeodomain protein PDX-1 exemplifies such interactions by binding to the A3/4 region of the rat insulin I promoter and activating insulin gene transcription by cooperating with the basic-helix-loop-helix (bHLH) protein E47/Pan1, which binds to the adjacent E2 site. The present study provides evidence that the homeodomain of PDX-1 acts as a protein-protein interaction domain to recruit multiple proteins, including E47/Pan1, BETA2/NeuroD1, and high-mobility group protein I(Y), to an activation complex on the E2A3/4 minienhancer. The transcriptional activity of this complex results from the clustering of multiple activation domains capable of interacting with coactivators and the basal transcriptional machinery. These interactions are not common to all homeodomain proteins: the LIM homeodomain protein Lmx1.1 can also activate the E2A3/4 minienhancer in cooperation with E47/Pan1 but does so through different interactions. Cooperation between Lmx1.1 and E47/Pan1 results not only in the aggregation of multiple activation domains but also in the unmasking of a potent activation domain on E47/Pan1 that is normally silent in non-beta cells. While more than one activation complex may be capable of activating insulin gene transcription through the E2A3/4 minienhancer, each is dependent on multiple specific interactions among a unique set of nuclear proteins.
Figures





References
-
- Abdulkadir S A, Casolaro V, Tai A K, Thanos D, Ono S J. High mobility group I/Y protein functions as a specific cofactor for Oct-2A: mapping of interaction domains. J Leukoc Biol. 1998;64:681–691. - PubMed
-
- Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. - PubMed
-
- Arlotta P, Rustighi A, Mantovani F, Manfioletti G, Giancotti V, Tell G, Damante G. High mobility group I proteins interfere with the homeodomains binding to DNA. J Biol Chem. 1997;272:29904–29910. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
