Exonic splicing enhancer motif recognized by human SC35 under splicing conditions
- PMID: 10629063
- PMCID: PMC85223
- DOI: 10.1128/MCB.20.3.1063-1071.2000
Exonic splicing enhancer motif recognized by human SC35 under splicing conditions
Abstract
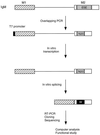
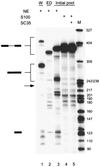
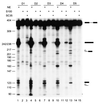
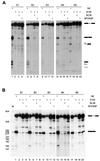
Exonic splicing enhancers (ESEs) are important cis elements required for exon inclusion. Using an in vitro functional selection and amplification procedure, we have identified a novel ESE motif recognized by the human SR protein SC35 under splicing conditions. The selected sequences are functional and specific: they promote splicing in nuclear extract or in S100 extract complemented by SC35 but not by SF2/ASF. They can also function in a different exonic context from the one used for the selection procedure. The selected sequences share one or two close matches to a short and highly degenerate octamer consensus, GRYYcSYR. A score matrix was generated from the selected sequences according to the nucleotide frequency at each position of their best match to the consensus motif. The SC35 score matrix, along with our previously reported SF2/ASF score matrix, was used to search the sequences of two well-characterized splicing substrates derived from the mouse immunoglobulin M (IgM) and human immunodeficiency virus tat genes. Multiple SC35 high-score motifs, but only two widely separated SF2/ASF motifs, were found in the IgM C4 exon, which can be spliced in S100 extract complemented by SC35. In contrast, multiple high-score motifs for both SF2/ASF and SC35 were found in a variant of the Tat T3 exon (lacking an SC35-specific silencer) whose splicing can be complemented by either SF2/ASF or SC35. The motif score matrix can help locate SC35-specific enhancers in natural exon sequences.
Figures


References
-
- Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. - PubMed
-
- Burge C B, Tuschl T, Sharp P A. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 525–559.
-
- Burset M, Guigo R. Evaluation of gene structure prediction programs. Genomics. 1996;34:353–367. - PubMed
-
- Cáceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
