Biochemical and molecular characterization of phenylacetate-coenzyme A ligase, an enzyme catalyzing the first step in aerobic metabolism of phenylacetic acid in Azoarcus evansii
- PMID: 10629172
- PMCID: PMC94275
- DOI: 10.1128/JB.182.2.286-294.2000
Biochemical and molecular characterization of phenylacetate-coenzyme A ligase, an enzyme catalyzing the first step in aerobic metabolism of phenylacetic acid in Azoarcus evansii
Abstract
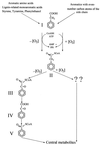
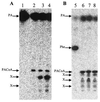
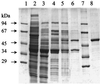
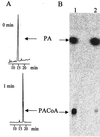
Phenylacetate-coenzyme A ligase (PA-CoA ligase; AMP forming, EC 6.2. 1.30), the enzyme catalyzing the first step in the aerobic degradation of phenylacetate (PA) in Azoarcus evansii, has been purified and characterized. The gene (paaK) coding for this enzyme was cloned and sequenced. The enzyme catalyzes the reaction of PA with CoA and MgATP to yield phenylacetyl-CoA (PACoA) plus AMP plus PPi. The enzyme was specifically induced after aerobic growth in a chemically defined medium containing PA or phenylalanine (Phe) as the sole carbon source. Growth with 4-hydroxyphenylacetate, benzoate, adipate, or acetate did not induce the synthesis of this enzyme. This enzymatic activity was detected very early in the exponential phase of growth, and a maximal specific activity of 76 nmol min(-1) mg of cell protein(-1) was measured. After 117-fold purification to homogeneity, a specific activity of 48 micromol min(-1) mg of protein(-1) was achieved with a turnover number (catalytic constant) of 40 s(-1). The protein is a monomer of 52 kDa and shows high specificity towards PA; other aromatic or aliphatic acids were not used as substrates. The apparent K(m) values for PA, ATP, and CoA were 14, 60, and 45 microM, respectively. The PA-CoA ligase has an optimum pH of 8 to 8.5 and a pI of 6.3. The enzyme is labile and requires the presence of glycerol for stabilization. The N-terminal amino acid sequence of the purified protein showed no homology with other reported PA-CoA ligases. The gene encoding this enzyme is 1, 320 bp long and codes for a protein of 48.75 kDa (440 amino acids) which shows high similarity with other reported PA-CoA ligases. An amino acid consensus for an AMP binding motif (VX2SSGTTGXP) was identified. The biochemical and molecular characteristics of this enzyme are quite different from those of the isoenzyme catalyzing the same reaction under anaerobic conditions in the same bacterium.
Figures

Similar articles
-
Molecular analysis of aerobic phenylacetate degradation in Azoarcus evansii.Mol Genet Genomics. 2002 Jul;267(5):656-63. doi: 10.1007/s00438-002-0699-9. Epub 2002 Jun 20. Mol Genet Genomics. 2002. PMID: 12172805
-
Reinvestigation of a new type of aerobic benzoate metabolism in the proteobacterium Azoarcus evansii.J Bacteriol. 2001 Mar;183(6):1899-908. doi: 10.1128/JB.183.6.1899-1908.2001. J Bacteriol. 2001. PMID: 11222587 Free PMC article.
-
Purification and characterization of phenylacetate-coenzyme A ligase from a denitrifying Pseudomonas sp., an enzyme involved in the anaerobic degradation of phenylacetate.Arch Microbiol. 1993;159(6):554-62. doi: 10.1007/BF00249035. Arch Microbiol. 1993. PMID: 8352645
-
Aryl Coenzyme A Ligases, a Subfamily of the Adenylate-Forming Enzyme Superfamily.Appl Environ Microbiol. 2021 Aug 26;87(18):e0069021. doi: 10.1128/AEM.00690-21. Epub 2021 Aug 26. Appl Environ Microbiol. 2021. PMID: 34260306 Free PMC article. Review.
-
Coenzyme A ligases involved in anaerobic biodegradation of aromatic compounds.Can J Microbiol. 1995 Oct;41(10):855-61. doi: 10.1139/m95-118. Can J Microbiol. 1995. PMID: 8590400 Review.
Cited by
-
Epoxy Coenzyme A Thioester pathways for degradation of aromatic compounds.Appl Environ Microbiol. 2012 Aug;78(15):5043-51. doi: 10.1128/AEM.00633-12. Epub 2012 May 11. Appl Environ Microbiol. 2012. PMID: 22582071 Free PMC article. Review.
-
Defining a structural and kinetic rationale for paralogous copies of phenylacetate-CoA ligases from the cystic fibrosis pathogen Burkholderia cenocepacia J2315.J Biol Chem. 2011 Apr 29;286(17):15577-85. doi: 10.1074/jbc.M111.219683. Epub 2011 Mar 8. J Biol Chem. 2011. PMID: 21388965 Free PMC article.
-
Structural Organization of Enzymes of the Phenylacetate Catabolic Hybrid Pathway.Biology (Basel). 2015 Jun 12;4(2):424-42. doi: 10.3390/biology4020424. Biology (Basel). 2015. PMID: 26075354 Free PMC article. Review.
-
Bacterial Tropone Natural Products and Derivatives: Overview of their Biosynthesis, Bioactivities, Ecological Role and Biotechnological Potential.Chembiochem. 2020 Sep 1;21(17):2384-2407. doi: 10.1002/cbic.201900786. Epub 2020 May 8. Chembiochem. 2020. PMID: 32239689 Free PMC article. Review.
-
Anaerobic catabolism of aromatic compounds: a genetic and genomic view.Microbiol Mol Biol Rev. 2009 Mar;73(1):71-133. doi: 10.1128/MMBR.00021-08. Microbiol Mol Biol Rev. 2009. PMID: 19258534 Free PMC article. Review.
References
-
- Altenschmidt U, Fuchs G. Novel aerobic 2-aminobenzoate metabolism. Purification and characterization of 2-aminobenzoate–CoA ligase, localization of the gene on a 8-kbp plasmid, and cloning and sequencing of the gene from a denitrifying Pseudomonas sp. Eur J Biochem. 1992;205:721–727. - PubMed
-
- Anders H-J, Kaetzke A, Kämpfer P, Ludwig W, Fuchs G. Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K172 and KB740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int J Syst Bacteriol. 1995;45:327–333. - PubMed
-
- Arunachalam U, Massey V, Vaidyanathan C S. p-Hydroxyphenylacetate-3-hydroxylase. A two-protein component enzyme. J Biol Chem. 1992;267:25848–25855. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

