Regulation of stalk elongation by phosphate in Caulobacter crescentus
- PMID: 10629178
- PMCID: PMC94281
- DOI: 10.1128/JB.182.2.337-347.2000
Regulation of stalk elongation by phosphate in Caulobacter crescentus
Abstract
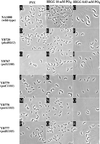
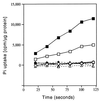
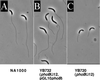
In Caulobacter crescentus, stalk biosynthesis is regulated by cell cycle cues and by extracellular phosphate concentration. Phosphate-starved cells undergo dramatic stalk elongation to produce stalks as much as 30 times as long as those of cells growing in phosphate-rich medium. To identify genes involved in the control of stalk elongation, transposon mutants were isolated that exhibited a long-stalk phenotype irrespective of extracellular phosphate concentration. The disrupted genes were identified as homologues of the high-affinity phosphate transport genes pstSCAB of Escherichia coli. In E. coli, pst mutants have a constitutively expressed phosphate (Pho) regulon. To determine if stalk elongation is regulated by the Pho regulon, the Caulobacter phoB gene that encodes the transcriptional activator of the Pho regulon was cloned and mutated. While phoB was not required for stalk synthesis or for the cell cycle timing of stalk synthesis initiation, it was required for stalk elongation in response to phosphate starvation. Both pstS and phoB mutants were deficient in phosphate transport. When a phoB mutant was grown with limiting phosphate concentrations, stalks only increased in length by an average of 1.4-fold compared to the average 9-fold increase in stalk length of wild-type cells grown in the same medium. Thus, the phenotypes of phoB and pst mutants were opposite. phoB mutants were unable to elongate stalks during phosphate starvation, whereas pst mutants made long stalks in both high- and low-phosphate media. Analysis of double pst phoB mutants indicated that the long-stalk phenotype of pst mutants was dependent on phoB. In addition, analysis of a pstS-lacZ transcriptional fusion showed that pstS transcription is dependent on phoB. These results suggest that the signal transduction pathway that stimulates stalk elongation in response to phosphate starvation is mediated by the Pst proteins and the response regulator PhoB.
Figures

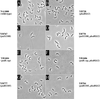
Similar articles
-
Identification of the PhoB Regulon and Role of PhoU in the Phosphate Starvation Response of Caulobacter crescentus.J Bacteriol. 2015 Oct 19;198(1):187-200. doi: 10.1128/JB.00658-15. Print 2016 Jan 1. J Bacteriol. 2015. PMID: 26483520 Free PMC article.
-
Sequential action of two-component genetic switches regulates the PHO regulon in Bacillus subtilis.J Bacteriol. 1994 Mar;176(5):1348-58. doi: 10.1128/jb.176.5.1348-1358.1994. J Bacteriol. 1994. PMID: 8113174 Free PMC article.
-
Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon.J Bacteriol. 1998 Mar;180(5):1277-86. doi: 10.1128/JB.180.5.1277-1286.1998. J Bacteriol. 1998. PMID: 9495769 Free PMC article.
-
Gene regulation by phosphate in enteric bacteria.J Cell Biochem. 1993 Jan;51(1):47-54. doi: 10.1002/jcb.240510110. J Cell Biochem. 1993. PMID: 8432742 Review.
-
From cell membrane to nucleotides: the phosphate regulon in Escherichia coli.Bioessays. 1990 Aug;12(8):371-6. doi: 10.1002/bies.950120804. Bioessays. 1990. PMID: 2241934 Review.
Cited by
-
The BAM complex subunit BamE (SmpA) is required for membrane integrity, stalk growth and normal levels of outer membrane {beta}-barrel proteins in Caulobacter crescentus.Microbiology (Reading). 2010 Mar;156(Pt 3):742-756. doi: 10.1099/mic.0.035055-0. Epub 2009 Dec 3. Microbiology (Reading). 2010. PMID: 19959579 Free PMC article.
-
Cell size control in bacteria.Curr Biol. 2012 May 8;22(9):R340-9. doi: 10.1016/j.cub.2012.02.032. Epub 2012 May 7. Curr Biol. 2012. PMID: 22575476 Free PMC article.
-
General protein diffusion barriers create compartments within bacterial cells.Cell. 2012 Dec 7;151(6):1270-82. doi: 10.1016/j.cell.2012.10.046. Epub 2012 Nov 29. Cell. 2012. PMID: 23201141 Free PMC article.
-
Regulation and properties of PstSCAB, a high-affinity, high-velocity phosphate transport system of Sinorhizobium meliloti.J Bacteriol. 2006 Feb;188(3):1089-102. doi: 10.1128/JB.188.3.1089-1102.2006. J Bacteriol. 2006. PMID: 16428413 Free PMC article.
-
Adhesion of single bacterial cells in the micronewton range.Proc Natl Acad Sci U S A. 2006 Apr 11;103(15):5764-8. doi: 10.1073/pnas.0601705103. Epub 2006 Apr 3. Proc Natl Acad Sci U S A. 2006. PMID: 16585522 Free PMC article.
References
-
- Aldridge P, Jenal U. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol Microbiol. 1999;32:379–392. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley/Greene; 1989.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

