Recombinational and mutational hotspots within the human lipoprotein lipase gene
- PMID: 10631137
- PMCID: PMC1288350
- DOI: 10.1086/302699
Recombinational and mutational hotspots within the human lipoprotein lipase gene
Abstract
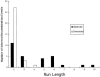
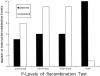
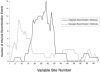
Here an analysis is presented of the roles of recombination and mutation in shaping previously determined haplotype variation in 9.7 kb of genomic DNA sequence from the human lipoprotein lipase gene (LPL), scored in 71 individuals from three populations: 24 African Americans, 24 Finns, and 23 non-Hispanic whites. Recombination and gene-conversion events inferred from data on 88 haplotypes that were defined by 69 variable sites were tested. The analysis revealed 29 statistically significant recombination events and one gene-conversion event. The recombination events were concentrated in a 1.9-kb region, near the middle of the segment, that contains a microsatellite and a pair of tandem and complementary mononucleotide runs; both the microsatellite and the runs show length variation. An analysis of site variation revealed that 9.6% of the nucleotides at CpG sites were variable, as were 3% of the nucleotides found in mononucleotide runs of >/=5 nucleotides, 3% of the nucleotides found </=3 bp from certain putative polymerase alpha-arrest sites, and 0. 5% of the remaining nucleotides. This nonhomogeneous distribution of variation suggests that multiple mutational hits at certain sites are common, an observation that challenges the fundamental assumption of the infinite-sites-mutation model. The nonrandom patterns of recombination and mutation suggest that randomly chosen single-nucleotide polymorphisms may not be optimal for disequilibrium mapping of this gene. Overall, these results indicate that both recombinational and mutational hotspots have played significant roles in shaping the haplotype variation at the LPL locus.
Figures
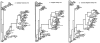
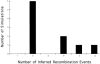

References
Electronic-Database Information
-
- GenBank, http://www.ncbi.nlm.nih.gov/Web/Genbank (for human LPL reference sequence [AF050163])
-
- MDECODE, http://mdecode.umich.edu/ (for a detailed view of the LPL statistical parsimony tree and for the details of the inferred recombination events)
References
-
- Carter C, Havlik R, Feinleib M, Kuller LH, Elston R, Rao DC (1984) Genetic epidemiology of coronary heart disease: past, present, and future. Arteriosclerosis 4:510–516 - PubMed
-
- Clark AG (1990) Inference of haplotypes from PCR-amplified samples of diploid populations. Mol Biol Evol 7:111–122 - PubMed
-
- Crandall KA (1994) Intraspecific cladogram estimation: accuracy at higher levels of divergence. Syst Biol 43:222–235
-
- Crandall KA, Templeton AR (1996) Applications of intraspecific phylogenetics. In: Harvey P, Brown AJL, Smith JM, Nee S (eds) New uses for new phylogenies. Oxford University Press, Oxford, pp 81–99
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

