High levels of sequence polymorphism and linkage disequilibrium at the telomere of 12q: implications for telomere biology and human evolution
- PMID: 10631154
- PMCID: PMC1288329
- DOI: 10.1086/302721
High levels of sequence polymorphism and linkage disequilibrium at the telomere of 12q: implications for telomere biology and human evolution
Abstract
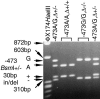
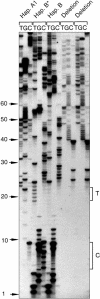
The human Xp/Yp telomere-junction region exhibits high levels of sequence polymorphism and linkage disequilibrium. To determine whether this is a general feature of human telomeres, we have undertaken sequence analysis at the 12q telomere and have extended the analysis at Xp/Yp. A total of 22 single-nucleotide polymorphisms (SNPs) and one 30-bp duplication were detected in the 1,870 bp adjacent to the 12q telomere. Twenty polymorphic positions were in almost complete linkage disequilibrium, creating three common diverged haplotypes accounting for 80% of 12q telomeres in the white population. A further 6% of 12q telomeres contained a 1,439-bp deletion in the DNA flanking the telomere. The remaining 13% of 12q telomeres did not amplify with the primers used (nulls). The distribution of telomere (TTAGGG) and variant repeats within 12q telomeres was hypervariable, but alleles with similar distribution patterns were associated with the same haplotype in the telomere-adjacent DNA. These data suggest that 12q telomeres, like Xp/Yp telomeres, exhibit low levels of homologous recombination and evolve along haploid lineages. In contrast, high levels of homologous recombination occur in the adjacent proterminal regions of human chromosomes. This suggests that there is a localized telomere-mediated suppression of recombination. In addition, the genetic characteristics of these regions may provide a source of deep lineages for the study of early human evolution, unaffected by both natural selection and recombination. To explain the presence of a few diverged haplotypes adjacent to the Xp/Yp and 12q telomeres, we propose a model that involves the hybridization of two archaic hominoid lineages ultimately giving rise to modern Homo sapiens.
Figures



References
Electronic-Database Information
-
- EMBL Database, http://helix.nih.gov/science/embl.html (for haplotype A [accession number AJ132885], haplotype A1 [accession number AJ132886], haplotype B [accession number AJ132887], and the sequence of the 12q deletion allele [accession number AJ132888])
-
- CEPH Genotype Database, http://www.cephb.fr/
References
-
- Baird DM, Royle NJ (1997) Sequences from higher primates orthologous to the human Xp/Yp telomere junction region reveal gross rearrangements and high levels of divergence. Hum Mol Genet 6:2291–2299 - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

