Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues
- PMID: 10631269
- PMCID: PMC58864
- DOI: 10.1104/pp.122.1.255
Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues
Abstract
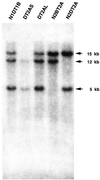
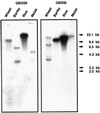
Studies of waxy mutations in wheat and other cereals have shown that null mutations in genes encoding granule-bound starch synthase I (GBSSI) result in amylose-free starch in endosperm and pollen grains, whereas starch in other tissues may contain amylose. We have isolated a cDNA from waxy wheat that encodes GBSSII, which is thought to be responsible for the elongation of amylose chains in non-storage tissues. The deduced amino acid sequences of wheat GBSSI and GBSSII were almost 66% identical, while those of wheat GBSSII and potato GBSSI were 72% identical. GBSSII was expressed in leaf, culm, and pericarp tissue, but transcripts were not detected in endosperm tissue. In contrast, GBSSI expression was high in endosperm tissue. The expression of GBSSII mRNA in pericarp tissue was similar at the midpoints of the day and night periods. The GBSSII genes were mapped to chromosomes 2AL, 2B, and 2D, whereas GBSSI genes are located on group 7 chromosomes. Gel-blot analysis indicated that genes related to GBSSII also occur in barley, rice, and maize. The possible role of GBSSII in starch synthesis is discussed.
Figures






Similar articles
-
Characterization of a granule-bound starch synthase isoform found in the pericarp of wheat.Plant Physiol. 1998 Oct;118(2):451-9. doi: 10.1104/pp.118.2.451. Plant Physiol. 1998. PMID: 9765530 Free PMC article.
-
The altered pattern of amylose accumulation in the endosperm of low-amylose barley cultivars is attributable to a single mutant allele of granule-bound starch synthase I with a deletion in the 5'-non-coding region.Plant Physiol. 2002 Sep;130(1):190-8. doi: 10.1104/pp.005454. Plant Physiol. 2002. PMID: 12226499 Free PMC article.
-
A 56-kDa protein is a novel granule-bound starch synthase existing in the pericarps, aleurone layers, and embryos of immature seed in diploid wheat (Triticum monococcum L.).Planta. 1998 Dec;207(1):125-32. doi: 10.1007/s004250050464. Planta. 1998. PMID: 9951718
-
Posttranslational Modification of Waxy to Genetically Improve Starch Quality in Rice Grain.Int J Mol Sci. 2021 May 3;22(9):4845. doi: 10.3390/ijms22094845. Int J Mol Sci. 2021. PMID: 34063649 Free PMC article. Review.
-
Granule-bound starch synthase in plants: Towards an understanding of their evolution, regulatory mechanisms, applications, and perspectives.Plant Sci. 2023 Nov;336:111843. doi: 10.1016/j.plantsci.2023.111843. Epub 2023 Aug 28. Plant Sci. 2023. PMID: 37648115 Review.
Cited by
-
Identification and testing of reference genes for Sesame gene expression analysis by quantitative real-time PCR.Planta. 2013 Mar;237(3):873-89. doi: 10.1007/s00425-012-1805-9. Epub 2012 Nov 16. Planta. 2013. PMID: 23229061 Free PMC article.
-
Improved 93-11 Genome and Time-Course Transcriptome Expand Resources for Rice Genomics.Front Plant Sci. 2022 Jan 21;12:769700. doi: 10.3389/fpls.2021.769700. eCollection 2021. Front Plant Sci. 2022. PMID: 35126409 Free PMC article.
-
Understanding Starch Metabolism in Pea Seeds towards Tailoring Functionality for Value-Added Utilization.Int J Mol Sci. 2021 Aug 20;22(16):8972. doi: 10.3390/ijms22168972. Int J Mol Sci. 2021. PMID: 34445676 Free PMC article. Review.
-
Transcriptomics analysis of hulless barley during grain development with a focus on starch biosynthesis.Funct Integr Genomics. 2017 Jan;17(1):107-117. doi: 10.1007/s10142-016-0537-5. Epub 2016 Dec 2. Funct Integr Genomics. 2017. PMID: 27913887 Free PMC article.
-
Multigene engineering of starch biosynthesis in maize endosperm increases the total starch content and the proportion of amylose.Transgenic Res. 2013 Dec;22(6):1133-42. doi: 10.1007/s11248-013-9717-4. Epub 2013 Jun 6. Transgenic Res. 2013. PMID: 23740205
References
-
- Abel GJW, Springer F, Willmitzer L, Kossmann J. Cloning and functional analysis of a cDNA encoding a novel 139 kDA starch synthase from potato (Solanum tuberosum L.) Plant J. 1996;10:981–991. - PubMed
-
- Ainsworth C, Clark J, Balsdon J. Expression, organisation and structure of the genes encoding the waxy protein (granule-bound starch synthase) in wheat. Plant Mol Biol. 1993a;22:67–82. - PubMed
-
- Ainsworth C, Tarvis M, Clark J. Isolation and analysis of a cDNA clone encoding the small subunit of ADP-glucose pyrophosphorylase from wheat. Plant Mol Biol. 1993b;23:22–33. - PubMed
-
- Badenhuizen NP. The Biosynthesis of Starch Granules in Higher Plants. New York: Appleton-Century-Crofts; 1969. pp. 1–115.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

