A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain
- PMID: 10632600
- PMCID: PMC6772408
- DOI: 10.1523/JNEUROSCI.20-02-00709.2000
A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain
Abstract
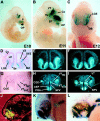
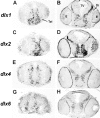
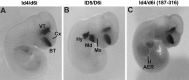
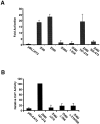
Four Dlx homeobox genes, Dlx1, Dlx2, Dlx5, and Dlx6 are expressed in the same primordia of the mouse forebrain with temporally overlapping patterns. The four genes are organized as two tail-to-tail pairs, Dlx1/Dlx2 and Dlx5/Dlx6, a genomic arrangement conserved in distantly related vertebrates like zebrafish. The Dlx5/Dlx6 intergenic region contains two sequences of a few hundred base pairs, remarkably well conserved between mouse and zebrafish. Reporter transgenes containing these two sequences are expressed in the forebrain of transgenic mice and zebrafish with patterns highly similar to endogenous Dlx5 and Dlx6 expression. The activity of the transgene is drastically reduced in mouse mutants lacking both Dlx1 and Dlx2, consistent with the decrease in endogenous Dlx5 and Dlx6 expression. These results suggest that cross-regulation by Dlx proteins, mediated by the intergenic sequences, is essential for Dlx5 and Dlx6 expression in the forebrain. This hypothesis is supported by cotransfection and DNA-protein binding experiments. We propose that the Dlx genes are part of a highly conserved developmental pathway that regulates forebrain development.
Figures




Similar articles
-
Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters.Genome Res. 2003 Apr;13(4):533-43. doi: 10.1101/gr.716103. Genome Res. 2003. PMID: 12670995 Free PMC article.
-
The proneural determinant MASH1 regulates forebrain Dlx1/2 expression through the I12b intergenic enhancer.Development. 2007 May;134(9):1755-65. doi: 10.1242/dev.02845. Epub 2007 Apr 4. Development. 2007. PMID: 17409112
-
Identification of a direct Dlx homeodomain target in the developing mouse forebrain and retina by optimization of chromatin immunoprecipitation.Nucleic Acids Res. 2004 Feb 9;32(3):884-92. doi: 10.1093/nar/gkh233. Print 2004. Nucleic Acids Res. 2004. PMID: 14769946 Free PMC article.
-
Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development.J Anat. 2005 Nov;207(5):501-61. doi: 10.1111/j.1469-7580.2005.00487.x. J Anat. 2005. PMID: 16313391 Free PMC article. Review.
-
Multiple functions of Dlx genes.Int J Dev Biol. 2000;44(6):619-26. Int J Dev Biol. 2000. PMID: 11061425 Review.
Cited by
-
Altered mRNA Splicing, Chondrocyte Gene Expression and Abnormal Skeletal Development due to SF3B4 Mutations in Rodriguez Acrofacial Dysostosis.PLoS Genet. 2016 Sep 13;12(9):e1006307. doi: 10.1371/journal.pgen.1006307. eCollection 2016 Sep. PLoS Genet. 2016. PMID: 27622494 Free PMC article.
-
Genetic approaches to elucidating cortical and hippocampal GABAergic interneuron diversity.Front Cell Neurosci. 2024 Jul 24;18:1414955. doi: 10.3389/fncel.2024.1414955. eCollection 2024. Front Cell Neurosci. 2024. PMID: 39113758 Free PMC article. Review.
-
Functional characterization of tissue-specific enhancers in the DLX5/6 locus.Hum Mol Genet. 2012 Nov 15;21(22):4930-8. doi: 10.1093/hmg/dds336. Epub 2012 Aug 21. Hum Mol Genet. 2012. PMID: 22914741 Free PMC article.
-
The role of gene duplication in the evolution and function of the vertebrate Dlx/distal-less bigene clusters.J Struct Funct Genomics. 2003;3(1-4):151-9. J Struct Funct Genomics. 2003. PMID: 12836694 Review.
-
Optogenetic Dissection of Neuronal Circuits in Zebrafish using Viral Gene Transfer and the Tet System.Front Neural Circuits. 2009 Dec 11;3:21. doi: 10.3389/neuro.04.021.2009. eCollection 2009. Front Neural Circuits. 2009. PMID: 20126518 Free PMC article.
References
-
- Acampora D, Merlo GR, Paleari L, Zeraga B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126:3795–3809. - PubMed
-
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JLR. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997a;278:474–476. - PubMed
-
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JLR. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late-born striatal neurons. Neuron. 1997b;19:27–37. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
