Identification and characterization of the nitrobenzene catabolic plasmids pNB1 and pNB2 in Pseudomonas putida HS12
- PMID: 10633088
- PMCID: PMC94317
- DOI: 10.1128/JB.182.3.573-580.2000
Identification and characterization of the nitrobenzene catabolic plasmids pNB1 and pNB2 in Pseudomonas putida HS12
Abstract
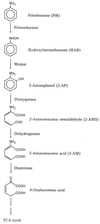
Pseudomonas putida HS12, which is able to grow on nitrobenzene, was found to carry two plasmids, pNB1 and pNB2. The activity assay experiments of wild-type HS12(pNB1 and pNB2), a spontaneous mutant HS121(pNB2), and a cured derivative HS124(pNB1) demonstrated that the catabolic genes coding for the nitrobenzene-degrading enzymes, designated nbz, are located on two plasmids, pNB1 and pNB2. The genes nbzA, nbzC, nbzD, and nbzE, encoding nitrobenzene nitroreductase, 2-aminophenol 1,6-dioxygenase, 2-aminomuconic 6-semialdehyde dehydrogenase, and 2-aminomuconate deaminase, respectively, are located on pNB1 (59.1 kb). Meanwhile, the nbzB gene encoding hydroxylaminobenzene mutase, a second-step enzyme in the nitrobenzene catabolic pathway, was found in pNB2 (43.8 kb). Physical mapping, cloning, and functional analysis of the two plasmids and their subclones in Escherichia coli strains revealed in more detail the genetic organization of the catabolic plasmids pNB1 and pNB2. The genes nbzA and nbzB are located on the 1.1-kb SmaI-SnaBI fragment of pNB1 and the 1.0-kb SspI-SphI fragment of pNB2, respectively, and their expressions were not tightly regulated. On the other hand, the genes nbzC, nbzD, and nbzE, involved in the ring cleavage pathway of 2-aminophenol, are localized on the 6.6-kb SnaBI-SmaI fragment of pNB1 and clustered in the order nbzC-nbzD-nbzE as an operon. The nbzCDE genes, which are transcribed in the opposite direction of the nbzA gene, are coordinately regulated by both nitrobenzene and a positive transcriptional regulator that seems to be encoded on pNB2.
Figures



Similar articles
-
Characterization of genes involved in the initial reactions of 4-chloronitrobenzene degradation in Pseudomonas putida ZWL73.Appl Microbiol Biotechnol. 2006 Nov;73(1):166-71. doi: 10.1007/s00253-006-0441-3. Epub 2006 Apr 27. Appl Microbiol Biotechnol. 2006. PMID: 16642329
-
p-Cumate catabolic pathway in Pseudomonas putida Fl: cloning and characterization of DNA carrying the cmt operon.J Bacteriol. 1996 Mar;178(5):1351-62. doi: 10.1128/jb.178.5.1351-1362.1996. J Bacteriol. 1996. PMID: 8631713 Free PMC article.
-
Implications of the xylQ gene of TOL plasmid pWW102 for the evolution of aromatic catabolic pathways.Microbiology (Reading). 1998 May;144 ( Pt 5):1387-1396. doi: 10.1099/00221287-144-5-1387. Microbiology (Reading). 1998. PMID: 9611813
-
Studies of the catabolic pathway of degradation of nitrobenzene by Pseudomonas pseudoalcaligenes JS45: removal of the amino group from 2-aminomuconic semialdehyde.Appl Environ Microbiol. 1997 Dec;63(12):4839-43. doi: 10.1128/aem.63.12.4839-4843.1997. Appl Environ Microbiol. 1997. PMID: 9471964 Free PMC article.
-
Genetic and structural organization of the aminophenol catabolic operon and its implication for evolutionary process.J Bacteriol. 2001 Sep;183(17):5074-81. doi: 10.1128/JB.183.17.5074-5081.2001. J Bacteriol. 2001. PMID: 11489860 Free PMC article.
Cited by
-
Biodegradation of high concentration of nitrobenzene by Pseudomonas corrugata embedded in peat-phosphate esterified polyvinyl alcohol.World J Microbiol Biotechnol. 2013 Oct;29(10):1859-67. doi: 10.1007/s11274-013-1348-7. Epub 2013 Apr 11. World J Microbiol Biotechnol. 2013. PMID: 23576015
-
A novel deaminase involved in chloronitrobenzene and nitrobenzene degradation with Comamonas sp. strain CNB-1.J Bacteriol. 2007 Apr;189(7):2677-82. doi: 10.1128/JB.01762-06. Epub 2007 Jan 26. J Bacteriol. 2007. PMID: 17259310 Free PMC article.
-
Integron diversity in heavy-metal-contaminated mine tailings and inferences about integron evolution.Appl Environ Microbiol. 2004 Feb;70(2):1160-8. doi: 10.1128/AEM.70.2.1160-1168.2004. Appl Environ Microbiol. 2004. PMID: 14766601 Free PMC article.
-
Nitroaromatic compounds, from synthesis to biodegradation.Microbiol Mol Biol Rev. 2010 Jun;74(2):250-72. doi: 10.1128/MMBR.00006-10. Microbiol Mol Biol Rev. 2010. PMID: 20508249 Free PMC article. Review.
-
Construction of Escherichia coli strains for conversion of nitroacetophenones to ortho-aminophenols.Appl Environ Microbiol. 2003 Nov;69(11):6520-6. doi: 10.1128/AEM.69.11.6520-6526.2003. Appl Environ Microbiol. 2003. PMID: 14602609 Free PMC article.
References
-
- Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

