Ralstonia eutropha TF93 is blocked in tat-mediated protein export
- PMID: 10633089
- PMCID: PMC94318
- DOI: 10.1128/JB.182.3.581-588.2000
Ralstonia eutropha TF93 is blocked in tat-mediated protein export
Abstract
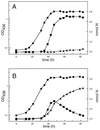
Ralstonia eutropha (formerly Alcaligenes eutrophus) TF93 is pleiotropically affected in the translocation of redox enzymes synthesized with an N-terminal signal peptide bearing a twin arginine (S/T-R-R-X-F-L-K) motif. Immunoblot analyses showed that the catalytic subunits of the membrane-bound [NiFe] hydrogenase (MBH) and the molybdenum cofactor-binding periplasmic nitrate reductase (Nap) are mislocalized to the cytoplasm and to the inner membrane, respectively. Moreover, physiological studies showed that the copper-containing nitrous oxide reductase (NosZ) was also not translocated to the periplasm in strain TF93. The cellular localization of enzymes exported by the general secretion system was unaffected. The translocation-arrested MBH and Nap proteins were enzymatically active, suggesting that twin-arginine signal peptide-dependent redox enzymes may have their cofactors inserted prior to transmembrane export. The periplasmic destination of MBH, Nap, and NosZ was restored by heterologous expression of Azotobacter chroococcum tatA mobilized into TF93. tatA encodes a bacterial Hcf106-like protein, a component of a novel protein transport system that has been characterized in thylakoids and shown to translocate folded proteins across the membrane.
Figures


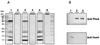


References
-
- Berks B C. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. - PubMed
-
- Bernhard M, Benelli B, Hochkoeppler A, Zannoni D, Friedrich B. Functional and structural role of the cytochrome b subunit of the membrane-bound hydrogenase complex of Alcaligenes eutrophus H16. Eur J Biochem. 1997;248:179–186. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

