Identification of an extradiol dioxygenase involved in tetralin biodegradation: gene sequence analysis and purification and characterization of the gene product
- PMID: 10633115
- PMCID: PMC94344
- DOI: 10.1128/JB.182.3.789-795.2000
Identification of an extradiol dioxygenase involved in tetralin biodegradation: gene sequence analysis and purification and characterization of the gene product
Abstract
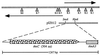
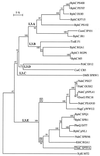
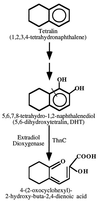
A genomic region involved in tetralin biodegradation was recently identified in Sphingomonas strain TFA. We have cloned and sequenced from this region a gene designated thnC, which codes for an extradiol dioxygenase required for tetralin utilization. Comparison to similar sequences allowed us to define a subfamily of 1, 2-dihydroxynaphthalene extradiol dioxygenases, which comprises two clearly different groups, and to show that ThnC clusters within group 2 of this subfamily. 1,2-Dihydroxy-5,6,7, 8-tetrahydronaphthalene was found to be the metabolite accumulated by a thnC insertion mutant. The ring cleavage product of this metabolite exhibited behavior typical of a hydroxymuconic semialdehyde toward pH-dependent changes and derivatization with ammonium to give a quinoline derivative. The gene product has been purified, and its biochemical properties have been studied. The enzyme is a decamer which requires Fe(II) for activity and shows high activity toward its substrate (V(max), 40.5 U mg(-1); K(m), 18. 6 microM). The enzyme shows even higher activity with 1, 2-dihydroxynaphthalene and also significant activity toward 1, 2-dihydroxybiphenyl or methylated catechols. The broad substrate specificity of ThnC is consistent with that exhibited by other extradiol dioxygenases of the same group within the subfamily of 1, 2-dihydroxynaphthalene dioxygenases.
Figures



References
-
- Asano Y, Yamamoto Y, Yamada H. Catechol 2,3-dioxygenase-catalyzed synthesis of picolinic acids from catechols. Biosci Biotech Biochem. 1994;58:2054–2056.
-
- Boldt Y, Whiting A, Wagner M, Sadowsky M, Que L, Jr, Wackett L. Manganese(II) active site mutants of 3,4-dihydroxyphenylacetate 2,3-dioxygenase from Arthrobacter globiformis strain CM-2. Biochemistry. 1997;36:2147–2153. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

