General properties and phylogenetic utilities of nuclear ribosomal DNA and mitochondrial DNA commonly used in molecular systematics
- PMID: 10634037
- PMCID: PMC2733198
- DOI: 10.3347/kjp.1999.37.4.215
General properties and phylogenetic utilities of nuclear ribosomal DNA and mitochondrial DNA commonly used in molecular systematics
Abstract
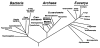
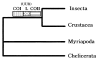
To choose one or more appropriate molecular markers or gene regions for resolving a particular systematic question among the organisms at a certain categorical level is still a very difficult process. The primary goal of this review, therefore, is to provide a theoretical information in choosing one or more molecular markers or gene regions by illustrating general properties and phylogenetic utilities of nuclear ribosomal DNA (rDNA) and mitochondrial DNA (mtDNA) that have been most commonly used for phylogenetic researches. The highly conserved molecular markers and/or gene regions are useful for investigating phylogenetic relationships at higher categorical levels (deep branches of evolutionary history). On the other hand, the hypervariable molecular markers and/or gene regions are useful for elucidating phylogenetic relationships at lower categorical levels (recently diverged branches). In summary, different selective forces have led to the evolution of various molecular markers or gene regions with varying degrees of sequence conservation. Thus, appropriate molecular markers or gene regions should be chosen with even greater caution to deduce true phylogenetic relationships over a broad taxonomic spectrum.
Figures







Similar articles
-
Arthropod phylogeny revisited, with a focus on crustacean relationships.Arthropod Struct Dev. 2010 Mar-May;39(2-3):88-110. doi: 10.1016/j.asd.2009.10.003. Epub 2009 Nov 5. Arthropod Struct Dev. 2010. PMID: 19854296
-
High 16S rDNA bacterial diversity in glacial meltwater lake sediment, Bratina Island, Antarctica.Extremophiles. 2003 Aug;7(4):275-82. doi: 10.1007/s00792-003-0321-z. Epub 2003 Apr 9. Extremophiles. 2003. PMID: 12910387
-
Testing the higher-level phylogenetic classification of Digenea (Platyhelminthes, Trematoda) based on nuclear rDNA sequences before entering the age of the 'next-generation' Tree of Life.J Helminthol. 2019 May;93(3):260-276. doi: 10.1017/S0022149X19000191. J Helminthol. 2019. PMID: 30973318
-
[Molecular markers and modern phylogenetics of mammals].Zh Obshch Biol. 2004 Jul-Aug;65(4):278-305. Zh Obshch Biol. 2004. PMID: 15490576 Review. Russian.
-
Nucleic acid techniques in bacterial systematics and identification.Int J Food Microbiol. 2007 Dec 15;120(3):225-36. doi: 10.1016/j.ijfoodmicro.2007.06.023. Epub 2007 Sep 5. Int J Food Microbiol. 2007. PMID: 17961780 Review.
Cited by
-
Identification and Phylogenetic Classification of Fasciola species Isolated from Sheep and Cattle by PCR-RFLP in Zabol, in Sistan and Baluchistan Province, Southeast Iran.Iran J Public Health. 2019 May;48(5):934-942. Iran J Public Health. 2019. PMID: 31523651 Free PMC article.
-
Liolophura species discrimination with geographical distribution patterns and their divergence and expansion history on the northwestern Pacific coast.Sci Rep. 2021 Sep 2;11(1):17602. doi: 10.1038/s41598-021-96823-5. Sci Rep. 2021. PMID: 34475451 Free PMC article.
-
Genetic variation of the freshwater snail Indoplanorbis exustus (Gastropoda: Planorbidae) in Thailand, inferred from 18S and 28S rDNA sequences.Parasitol Res. 2024 Jan 12;123(1):93. doi: 10.1007/s00436-024-08120-5. Parasitol Res. 2024. PMID: 38212518
-
Genetic characterization of the causative agent of besnoitiosis in goats in Iran on the basis of internal transcribed spacer rDNA and its comparison with Besnoitia species of other hosts.Parasitol Res. 2011 Mar;108(3):633-8. doi: 10.1007/s00436-010-2107-4. Epub 2010 Oct 13. Parasitol Res. 2011. PMID: 20941630
-
Mitochondrial DNA of Sardinian and North-West Italian Populations Revealed a New Piece in the Mosaic of Phylogeography and Phylogeny of Salariopsis fluviatilis (Blenniidae).Animals (Basel). 2022 Dec 2;12(23):3403. doi: 10.3390/ani12233403. Animals (Basel). 2022. PMID: 36496923 Free PMC article.
References
-
- Abele LG, Kim W, Felegenhauer BE. Molecular evidence for inclusion of the phylum Pentastomida in the Crustacea. Mol Biol Evol. 1989;6:685–691.
-
- Aguinaldo AMA, Turbeville JM, Linford LS, et al. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–493. - PubMed
-
- Bachellerie JP, Michot B. Evolution of large subunit rRNA structure. The 3' terminal domain contains elements of secondary structure specific to major phylogenetic groups. Biochimie. 1989;71:701–709. - PubMed
-
- Ballard JWO, Olsen GJ, Faith DP, Odgers WA, Rowell DM, Atkinson PW. Evidence from 12S ribosomal RNA sequences that onychophorans are modified arthropods. Science. 1992;258:1345–1348. - PubMed
-
- Beaver PC, Jung RC, Cupp EW. Clinical parasitology. 9th ed. Philadelphia, USA: Lea & Febiger; 1984.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

