Overexpression, purification, and partial characterization of ADP-ribosyltransferases modA and modB of bacteriophage T4
- PMID: 10634320
- PMCID: PMC6157369
Overexpression, purification, and partial characterization of ADP-ribosyltransferases modA and modB of bacteriophage T4
Abstract
There is increasing experimental evidence that ADP-ribosylation of host proteins is an important means to regulate gene expression of bacteriophage T4. Surprisingly, this phage codes for three different ADP-ribosyltransferases, gene products Alt, ModA, and ModB, modifying partially overlapping sets of host proteins. While gene product Alt already has been isolated as a recombinant protein and its action on host RNA polymerases and transcription regulation have been studied, the nucleotide sequences of the two mod genes was published only recently. Their mode of action in the course of the infection cycle and the consequences of the ADP-ribosylations catalyzed by these enzymes remain to be investigated. Here we describe the cloning of the genes, the overexpression, purification, and partial characterization of ADP-ribosyltransferases ModA and ModB. Both proteins seem to act independently, and the ADP-ribosyl moieties are transferred to different sets of host proteins. While gene product ModA, similarly to the Alt protein, acts also on the alpha-subunit of host RNA polymerase, the ModB activity serves another set of proteins, one of which was identified as the S1 protein associated with the 30S subunit of the E. coli ribosomes.
Figures

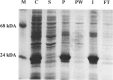

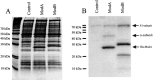

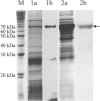
References
-
- Ausubel F. M.; Brent R.; Kingston R. E.; Moore D. D.; Seidman J. G.; Smith J. A.; Struhl K. Current protocols in molecular biology. In: Anonymous current protocols in molecular biology. New York: Greene Publishing and Wiley-Interscience; 1993.
-
- Bazan J. F.; Koch-Nolte F. Sequence and structural links between distant ADP-ribosyltransferase families. Adv. Exp. Med. Biol. 419:99–107; 1997. - PubMed
-
- Dagert M.; Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene 6:23–28; 1979. - PubMed
-
- Edman P. Sequence determination. Mol. Biol. Biochem. Biophys. 8:211–255; 1970. - PubMed
-
- Frazier M. W.; Mosig G. The bacteriophage T4 gene mrh whose product inhibits late T4 gene expression in an Escherichia coli rpoH (sigma 32) mutant. Gene 88: 7–14; 1990. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
