Determining the physical limits of the Brassica S locus by recombinational analysis
- PMID: 10634905
- PMCID: PMC140212
- DOI: 10.1105/tpc.12.1.23
Determining the physical limits of the Brassica S locus by recombinational analysis
Abstract
A genetic analysis was performed to study the frequency of recombination for intervals across the Brassica S locus region. No recombination was observed between the S locus glycoprotein gene and the S receptor kinase gene in the segregating populations that we analyzed. However, a number of recombination breakpoints in regions flanking these genes were identified, allowing the construction of an integrated genetic and physical map of the genomic region encompassing one S haplotype. We identified, based on the pollination phenotype of plants homozygous for recombinant S haplotypes, a 50-kb region that encompasses all specificity functions in the S haplotype that we analyzed. Mechanisms that might operate to preserve the tight linkage of self-incompatibility specificity genes within the S locus complex are discussed in light of the relatively uniform recombination frequencies that we observed across the S locus region and of the structural heteromorphisms that characterize different S haplotypes.
Figures



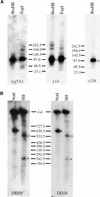

References
-
- Boyes, D.C., and Nasrallah, J.B. (1993). Physical linkage of the SLG and SRK genes at the self-incompatibility locus of Brassica oleracea. Mol. Gen. Genet. 236 369–373. - PubMed
-
- Camargo, L.E.A., Savides, L., Jung, G., Nienhuis, J., and Osborn, T.C. (1997). Location of the self-incompatibility locus in an RFLP and RAPD map of Brassica oleracea. J. Hered. 88 57–60. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

