A membrane-embedded glutamate is required for ligand binding to the multidrug transporter EmrE
- PMID: 10637227
- PMCID: PMC305557
- DOI: 10.1093/emboj/19.2.234
A membrane-embedded glutamate is required for ligand binding to the multidrug transporter EmrE
Abstract
EmrE is an Escherichia coli multidrug transporter that confers resistance to a variety of toxins by removing them in exchange for hydrogen ions. The detergent-solubilized protein binds tetraphenylphosphonium (TPP(+)) with a K(D) of 10 nM. One mole of ligand is bound per approximately 3 mol of EmrE, suggesting that there is one binding site per trimer. The steep pH dependence of binding suggests that one or more residues, with an apparent pK of approximately 7.5, release protons prior to ligand binding. A conservative Asp replacement (E14D) at position 14 of the only membrane-embedded charged residue shows little transport activity, but binds TPP(+) at levels similar to those of the wild-type protein. The apparent pK of the Asp shifts to <5.0. The data are consistent with a mechanism requiring Glu14 for both substrate and proton recognition. We propose a model in which two of the three Glu14s in the postulated trimeric EmrE homooligomer deprotonate upon ligand binding. The ligand is released on the other face of the membrane after binding of protons to Glu14.
Figures
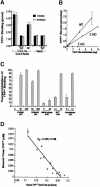

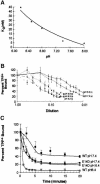
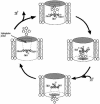
References
-
- Arkin I., Russ, W., Lebendiker, M. and Schuldiner, S. (1996) Determining the secondary structure and orientation of EmrE, a multi-drug transporter, indicates a transmembrane four helix bundle. Biochemistry, 35, 7233–7238. - PubMed
-
- Gottesman M.M., Pastan, I. and Ambudkar, S.V. (1996) P-glycoprotein and multidrug resistance. Curr. Opin. Genet. Dev., 6, 610–617. - PubMed
-
- Grinius L. and Goldberg, E. (1994) Bacterial multidrug resistance is due to a single membrane protein which functions as a drug pump. J. Biol. Chem., 269, 29998–30004. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

