ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking
- PMID: 10637304
- PMCID: PMC14770
- DOI: 10.1091/mbc.11.1.227
ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking
Abstract
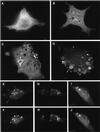
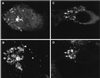
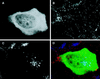
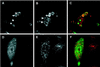
The yeast vacuolar sorting protein Vps4p is an ATPase required for endosomal trafficking that couples membrane association to its ATPase cycle. To investigate the function of mammalian VPS4 in endosomal trafficking, we have transiently expressed wild-type or ATPase-defective human VPS4 (hVPS4) in cultured cells. Wild-type hVPS4 was cytosolic, whereas a substantial fraction of hVPS4 that was unable to either bind or hydrolyze ATP was localized to membranes, including those of specifically induced vacuoles. Vacuoles were exclusively endocytic in origin, and subsets of enlarged vacuoles stained with markers for each stage of the endocytic pathway. Sorting of receptors from the early endosome to the recycling compartment or to the trans-Golgi network was not significantly affected, and no mutant hVPS4 associated with these compartments. However, many hVPS4-induced vacuoles were substantially enriched in cholesterol relative to the endosomal compartments of untransfected cells, indicating that expression of mutant hVPS4 gives rise to a kinetic block in postendosomal cholesterol sorting. The phenotype described here is largely consistent with the defects in vacuolar sorting associated with class E vps mutants in yeast, and a role for mammalian VPS4 is discussed in this context.
Figures




References
-
- Adams MD, Dubnick M, Kerlavage AR, Moreno R, Kelley JM, Utterback TR, Nagle JW, Fields C, Venter JC. Sequence identification of 2375 human brain genes. Nature. 1992;355:632–634. - PubMed
-
- Bright NA, Reaves BJ, Mullock BM, Luzio JP. Dense core lysosomes can fuse with late endosomes and are reformed from the resultant hybrid organelles. J Cell Sci. 1997;110:2027–2040. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

