Optimised ligation of oligonucleotides by thermal ligases: comparison of Thermus scotoductus and Rhodothermus marinus DNA ligases to other thermophilic ligases
- PMID: 10637340
- PMCID: PMC102565
- DOI: 10.1093/nar/28.3.e10
Optimised ligation of oligonucleotides by thermal ligases: comparison of Thermus scotoductus and Rhodothermus marinus DNA ligases to other thermophilic ligases
Abstract
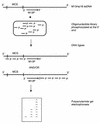
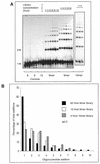
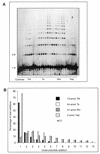
We describe the characterisation of four thermo-stable NAD(+)-dependent DNA ligases, from Thermus thermophilus (Tth), Thermus scotoductus (Ts), Rhodothermus marinus (Rm) and Thermus aquaticus (Taq), by an assay which measures ligation rate and mismatch discrimination. Complete libraries of octa-, nona- and decanucleotides were used as substrates. The assay comprised the polymerisation of oligo-nucleotides initiated from a 17 base 'primer', using M13mp18 ssDNA as template. Polymers of ligation products were analysed by polyacrylamide gel electro-phoresis. Under optimum conditions, the enzymes produced polymers ranging from 8 to 16 additions; there was variation between enzymes and the length of the oligonucleotides had a strong effect. The optimal total oligonucleotide concentration for each library was approximately 4 nmol. We compared the rates of ligation between the four ligases using an octanucleotide library as substrate. By this criterion, the Ts and Rm ligases are far more active compared to the more commonly available thermostable ligases.
Figures
References
-
- Waga S. and Stillman,B. (1998) Annu. Rev. Biochem., 67, 721–751. - PubMed
-
- Tomkinson A.E. and Levin,D.S. (1997) Bioessays, 19, 893–901. - PubMed
-
- Landegren U., Kaiser,R., Sanders,J. and Hood,L. (1988) Science, 241, 1077–1080. - PubMed
-
- Baron H., Fung,S., Aydin,A., Bahring,S., Luft,F.C. and Schuster,H. (1996) Nature Biotechnol., 14, 1279–1282. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

