Identification of the operon for the sorbitol (Glucitol) Phosphoenolpyruvate:Sugar phosphotransferase system in Streptococcus mutans
- PMID: 10639465
- PMCID: PMC97224
- DOI: 10.1128/IAI.68.2.925-930.2000
Identification of the operon for the sorbitol (Glucitol) Phosphoenolpyruvate:Sugar phosphotransferase system in Streptococcus mutans
Abstract
Transposon mutagenesis and marker rescue were used to isolate and identify an 8.5-kb contiguous region containing six open reading frames constituting the operon for the sorbitol P-enolpyruvate phosphotransferase transport system (PTS) of Streptococcus mutans LT11. The first gene, srlD, codes for sorbitol-6-phosphate dehydrogenase, followed downstream by srlR, coding for a transcriptional regulator; srlM, coding for a putative activator; and the srlA, srlE, and srlB genes, coding for the EIIC, EIIBC, and EIIA components of the sorbitol PTS, respectively. Among all sorbitol PTS operons characterized to date, the srlD gene is found after the genes coding for the EII components; thus, the location of the gene in S. mutans is unique. The SrlR protein is similar to several transcriptional regulators found in Bacillus spp. that contain PTS regulator domains (J. Stülke, M. Arnaud, G. Rapoport, and I. Martin-Verstraete, Mol. Microbiol. 28:865-874, 1998), and its gene overlaps the srlM gene by 1 bp. The arrangement of these two regulatory genes is unique, having not been reported for other bacteria.
Figures



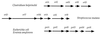
References
-
- Aldridge P, Metzger M, Geider K. Genetics of sorbitol metabolism in Erwinia amylovora and its influence on bacterial virulence. Mol Gen Genet. 1997;256:611–619. - PubMed
-
- Birkhed D, Edwardsson S, Kalfas S, Svensäter G. Cariogenicity of sorbitol. Swed Dent J. 1984;8:147–154. - PubMed
-
- Bowden G H W. Which bacteria are cariogenic in humans? In: Johnson N M, editor. Dental caries. 1. Markers of high and low risk groups and individuals. Cambridge, United Kingdom: Cambridge University Press; 1991. pp. 266–286.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

