Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus Avian infectious bronchitis virus and characterization of the cleavage products
- PMID: 10644337
- PMCID: PMC111642
- DOI: 10.1128/jvi.74.4.1674-1685.2000
Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus Avian infectious bronchitis virus and characterization of the cleavage products
Abstract
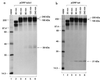
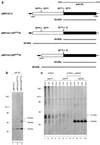
The coronavirus Avian infectious bronchitis virus (IBV) employs polyprotein processing as a strategy to express its gene products. Previously we identified the first cleavage event as proteolysis at the Gly(673)-Gly(674) dipeptide bond mediated by the first papain-like proteinase domain (PLPD-1) to release an 87-kDa mature protein. In this report, we demonstrate a novel cleavage activity of PLPD-1. Expression, deletion, and mutagenesis studies showed that the product encoded between nucleotides 2548 and 8865 was further cleaved by PLPD-1 at the Gly(2265)-Gly(2266) dipeptide bond to release an N-terminal 195-kDa and a C-terminal 41-kDa cleavage product. Characterization of the cleavage activity revealed that the proteinase is active on this scissile bond when expressed in vitro in rabbit reticulocyte lysates and can act on the same substrate in trans when expressed in intact cells. Both the N- and C-terminal cleavage products were detected in virus-infected cells and were found to be physically associated. Glycosidase digestion and site-directed mutagenesis studies of the 41-kDa protein demonstrated that it is modified by N-linked glycosylation at the Asn(2313) residue encoded by nucleotides 7465 to 7467. By using a region-specific antiserum raised against the IBV sequence encoded by nucleotides 8865 to 9786, we also demonstrated that a 33-kDa protein, representing the 3C-like proteinase (3CLP), was specifically immunoprecipitated from the virus-infected cells. Site-directed mutagenesis and expression studies showed that a previously predicted cleavage site (Q(2583)-G(2584)) located within the 41-kDa protein-encoding region was not utilized by 3CLP, supporting the conclusion that the 41-kDa protein is a mature viral product.
Figures








References
-
- Boursnell M E G, Brown T D K, Foulds I J, Green P F, Tomley F M, Binns M M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987;68:57–77. - PubMed
-
- Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutant of the green fluorescent protein (GFP) Gene. 1996;173:33–38. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

