The central proline of an internal viral fusion peptide serves two important roles
- PMID: 10644338
- PMCID: PMC111643
- DOI: 10.1128/jvi.74.4.1686-1693.2000
The central proline of an internal viral fusion peptide serves two important roles
Abstract
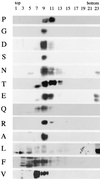
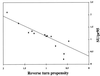
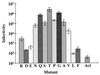
The fusion peptide of the avian sarcoma/leukosis virus (ASLV) envelope protein (Env) is internal, near the N terminus of its transmembrane (TM) subunit. As for most internal viral fusion peptides, there is a proline near the center of this sequence. Robson-Garnier structure predictions of the ASLV fusion peptide and immediate surrounding sequences indicate a region of order (beta-sheet), a tight reverse turn containing the proline, and a second region of order (alpha-helix). Similar motifs (order, turn or loop, order) are predicted for other internal fusion peptides. In this study, we made and analyzed 12 Env proteins with substitutions for the central proline of the fusion peptide. Env proteins were expressed in 293T cells and in murine leukemia virus pseudotyped virions. We found the following. (i) All mutant Envs form trimers, but when the bulky hydrophobic residues phenylalanine or leucine are substituted for proline, trimerization is weakened. (ii) Surprisingly, the proline is required for maximal processing of the Env precursor into its surface and TM subunits; the amount of processing correlates linearly with the propensity of the substituted residue to be found in a reverse turn. (iii) Nonetheless, proteolytically processed forms of all Envs are preferentially incorporated into pseudotyped virions. (iv) All Envs bind receptor with affinity greater than or equal to wild-type affinity. (v) Residues that support high infectivity cluster with proline at intermediate hydrophobicity. Infectivity is not supported by mutant Envs in which charged residues are substituted for proline, nor is it supported by the trimerization-defective phenylalanine and leucine mutants. Our findings suggest that the central proline in the ASLV fusion peptide is important for the formation of the native (metastable) Env structure as well as for membrane interactions that lead to fusion.
Figures




References
-
- Baker K A, Dutch R E, Lamb R A, Jardetzky T S. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell. 1999;3:309–319. - PubMed
-
- Bigler D, Chen M, Waters S, White J M. A model for sperm-egg binding and fusion based on ADAMs and integrins. Trends Cell Biol. 1997;7:220–225. - PubMed
-
- Blobel C P, Wolfsberg T G, Turck C W, Myles D G, Primakoff P, White J M. A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature. 1992;356:248–252. - PubMed
-
- Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

