Multiple integrations of human foamy virus in persistently infected human erythroleukemia cells
- PMID: 10644342
- PMCID: PMC111647
- DOI: 10.1128/jvi.74.4.1718-1726.2000
Multiple integrations of human foamy virus in persistently infected human erythroleukemia cells
Abstract
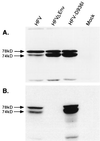
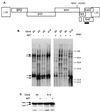
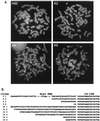
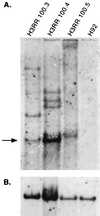
Foamy viruses are complex retroviruses whose replication strategy resembles that of conventional retroviruses. However, foamy virus replication also resembles that of hepadnaviruses in many respects. Because hepadnaviruses replicate in an integrase-independent manner, we were interested in investigating the characteristics of human foamy virus (HFV) integration. We have shown that HFV requires a functional integrase protein for infectivity. Our analyses have revealed that in single-cell clones derived from HFV-infected erythroleukemia-derived cells (H92), there were up to 20 proviral copies per host cell genome as determined by Southern blot and fluorescent in situ hybridization analysis. Use of specific probes has also shown that a majority of the proviruses contain the complete tas gene, which encodes the viral transactivator, and are not derived from Deltatas cDNAs, which have been shown to arise rapidly in infected cells. To demonstrate that the multiple proviral sequences are due to integration instead of recombination, we have sequenced the junctions between the proviral sequences and the host genome and found that the proviruses have authentic long terminal repeat ends and that each integration is at a different chromosomal site. A virus lacking the Gag nuclear localization signal accumulates fewer proviruses, suggesting that nuclear translocation is important for high proviral load. Since persistently infected H92 clones are not resistant to superinfection, the relative importance of an intracellular versus extracellular mechanism in proviral acquisition has yet to be determined.
Figures



References
-
- Achong B G, Mansell P W A, Epstein M A, Clifford P. An unusual virus in cultures from a human nasopharyngeal carcinoma. J Natl Cancer Inst. 1971;46:299–307. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1998.
-
- Birren B W, Tachi-iri Y, Kim U J, Nguyen M, Shizuya H, Korenberg J R, Simon M I. A human chromosome 22 fosmid resource: mapping and analysis of 96 clones. Genomics. 1996;34:97–106. - PubMed
-
- Bock C T, Schwinn S, Schroder C H, Velhagen I, Zentgraf H. Localization of hepatitis B virus core protein and viral DNA at the nuclear membrane. Virus Genes. 1996;12:53–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

