Disruption of the murine gammaherpesvirus 68 M1 open reading frame leads to enhanced reactivation from latency
- PMID: 10644370
- PMCID: PMC111675
- DOI: 10.1128/jvi.74.4.1973-1984.2000
Disruption of the murine gammaherpesvirus 68 M1 open reading frame leads to enhanced reactivation from latency
Abstract
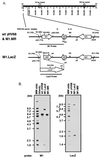
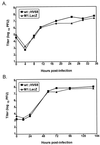
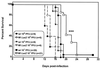
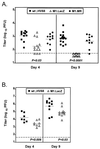
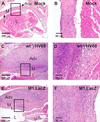
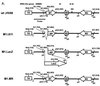
Murine gammaherpesvirus 68 (gammaHV68, or MHV-68) is a genetically tractable, small animal model for the analysis of gammaherpesvirus pathogenesis. The gammaHV68 genome is colinear with the genomes of other sequence gammaherpesviruses, containing large blocks of conserved genes interspersed by a number of putative genes without clear homologs in the other gammaherpesviruses. One of these putative unique genes, the M1 open reading frame (ORF), exhibits sequence homology to a poxvirus serine protease inhibitor, SPI-1, as well as to another gammaHV68 gene, M3, which we have recently shown encodes an abundantly secreted chemokine binding protein. To assess the contribution of the M1 ORF to gammaHV68 pathogenesis, we have generated a recombinant gammaHV68 in which the M1 ORF has been disrupted through targeted insertion of a lacZ expression cassette (M1.LacZ). Although M1.LacZ replicated normally in tissue culture, it exhibited decreased splenic titers at days 4 and 9 postinfection in both immunocompetent and immunodeficient mice. Despite decreased levels of acute virus replication, M1.LacZ established a latent infection comparable to wild-type (wt) gammaHV68, but exhibited an approximately fivefold increase in efficiency of reactivation from latency. M1.LacZ also caused severe vasculitis of the great elastic arteries in gamma interferon receptor (IFN-gammaR)-deficient mice with a frequency comparable to wt gammaHV68, but did not cause the mortality or splenic pathology observed with wt gammaHV68 infection of IFN-gammaR-deficient mice. Restoration of M1 ORF sequences into M1.LacZ (M1 marker rescue, or M1.MR) demonstrated that M1.LacZ phenotypic alterations in growth in vivo and latency were not due to the presence of additional mutations located elsewhere in the M1. LacZ genome. Generation of a second M1 mutant virus containing a deletion at the 5' end of the M1 ORF (M1Delta511), but lacking the LacZ expression cassette, revealed the same latency phenotype observed with the M1.LacZ mutant. However, M1Delta511 was not attenuated for acute virus replication in the spleen. We conclude that (i) the induction of arteritis in gammaHV68-infected IFN-gammaR-deficient mice can occur in the absence of splenic pathology and mortality, (ii) replication during acute infection is not the primary determinant for the establishment of latent infection, and (iii) the M1 ORF, or a closely linked gene, encodes a gene product that functions to suppress virus reactivation.
Figures






References
-
- Ali A N, Turner P C, Brooks M A, Moyer R W. The SPI-1 gene of rabbitpox virus determines host range and is required for hemorrhagic pock formation. Virology. 1994;202:305–314. - PubMed
-
- Bosma M J. The scid mutation: occurrence and effect. Curr Top Microbiol Immunol. 1989;152:3–9. - PubMed
-
- Bowden R J, Simas J P, Davis A J, Efstathiou S. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J Gen Virol. 1997;78:1675–1687. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

