Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA
- PMID: 10648561
- PMCID: PMC2174275
- DOI: 10.1083/jcb.148.2.283
Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA
Abstract
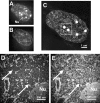
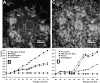
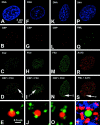
The promyelocytic leukemia (PML) nuclear body (also referred to as ND10, POD, and Kr body) is involved in oncogenesis and viral infection. This subnuclear domain has been reported to be rich in RNA and a site of nascent RNA synthesis, implicating its direct involvement in the regulation of gene expression. We used an analytical transmission electron microscopic method to determine the structure and composition of PML nuclear bodies and the surrounding nucleoplasm. Electron spectroscopic imaging (ESI) demonstrates that the core of the PML nuclear body is a dense, protein-based structure, 250 nm in diameter, which does not contain detectable nucleic acid. Although PML nuclear bodies contain neither chromatin nor nascent RNA, newly synthesized RNA is associated with the periphery of the PML nuclear body, and is found within the chromatin-depleted region of the nucleoplasm immediately surrounding the core of the PML nuclear body. We further show that the RNA does not accumulate in the protein core of the structure. Our results dismiss the hypothesis that the PML nuclear body is a site of transcription, but support the model in which the PML nuclear body may contribute to the formation of a favorable nuclear environment for the expression of specific genes.
Figures



References
-
- Bazett-Jones D.P., Hendzel M.J. Electron spectroscopic imaging of chromatin. Companion Meth. Enzymol. 1999;17:188–200 . - PubMed
-
- Bazett-Jones D.P., Hendzel M.J., Kruhlak M.J. Stoichiometric analysis of protein- and nucleic acid-based structures in the cell nucleus. Micron. 1999;30:151–157 . - PubMed
-
- Bernhard W. A new staining procedure for electron microscopical cytology. J. Ultrastruct. Res. 1969;27:250–265 . - PubMed
-
- Boggs B.A., Connors B., Sobel R.E., Chinaud A.C., Allis C.D. Reduced levels of histone H3 acetylation on the inactive X chromosomes as shown by histone acetylation. Chromosoma. 1996;105:41–49 . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

