Kinetics of ribosomal pausing during programmed -1 translational frameshifting
- PMID: 10648594
- PMCID: PMC85227
- DOI: 10.1128/MCB.20.4.1095-1103.2000
Kinetics of ribosomal pausing during programmed -1 translational frameshifting
Abstract
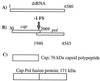
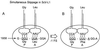
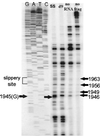
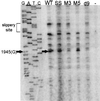
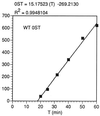
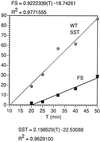
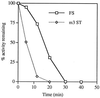
In the Saccharomyces cerevisiae double-stranded RNA virus, programmed -1 ribosomal frameshifting is responsible for translation of the second open reading frame of the essential viral RNA. A typical slippery site and downstream pseudoknot are necessary for this frameshifting event, and previous work has demonstrated that ribosomes pause over the slippery site. The translational intermediate associated with a ribosome paused at this position is detected, and, using in vitro translation and quantitative heelprinting, the rates of synthesis, the ribosomal pause time, the proportion of ribosomes paused at the slippery site, and the fraction of paused ribosomes that frameshift are estimated. About 10% of ribosomes pause at the slippery site in vitro, and some 60% of these continue in the -1 frame. Ribosomes that continue in the -1 frame pause about 10 times longer than it takes to complete a peptide bond in vitro. Altering the rate of translational initiation alters the rate of frameshifting in vivo. Our in vitro and in vivo experiments can best be interpreted to mean that there are three methods by which ribosomes pass the frameshift site, only one of which results in frameshifting.
Figures
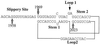
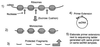



References
-
- Anderson R P, Menninger J R. Tests of the ribosome editor hypothesis. III. A mutant Escherichia coli with a defective ribosome editor. Mol Gen Genet. 1987;209:313–318. - PubMed
-
- Bostian K A, Elliott Q, Bussey H, Burn V, Smith A, Tipper D J. Sequence of the preprotoxin dsRNA gene of type 1 killer yeast: multiple processing events produce a two-component toxin. Cell. 1984;36:741–751. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
