RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates
- PMID: 10648605
- PMCID: PMC85244
- DOI: 10.1128/MCB.20.4.1194-1205.2000
RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates
Abstract
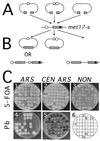
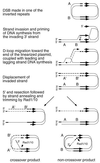
DNA double-strand breaks may be induced by endonucleases, ionizing radiation, chemical agents, and mechanical forces or by replication of single-stranded nicked chromosomes. Repair of double-strand breaks can occur by homologous recombination or by nonhomologous end joining. A system was developed to measure the efficiency of plasmid gap repair by homologous recombination using either chromosomal or plasmid templates. Gap repair was biased toward gene conversion events unassociated with crossing over using either donor sequence. The dependence of recombinational gap repair on genes belonging to the RAD52 epistasis group was tested in this system. RAD51, RAD52, RAD57, and RAD59 were required for efficient gap repair using either chromosomal or plasmid donors. No homologous recombination products were recovered from rad52 mutants, whereas a low level of repair occurred in the absence of RAD51, RAD57, or RAD59. These results suggest a minor pathway of strand invasion that is dependent on RAD52 but not on RAD51. The residual repair events in rad51 mutants were more frequently associated with crossing over than was observed in the wild-type strain, suggesting that the mechanisms for RAD51-dependent and RAD51-independent events are different. Plasmid gap repair was reduced synergistically in rad51 rad59 double mutants, indicating an important role for RAD59 in RAD51-independent repair.
Figures



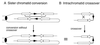
References
-
- Aguilera A. Genetic evidence for different RAD52-dependent intrachromosomal recombination pathways in Saccharomyces cerevisiae. Curr Genet. 1995;27:298–305. - PubMed
-
- Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. - PubMed
-
- Bai Y, Symington L S. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
