Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9
- PMID: 10648611
- PMCID: PMC85258
- DOI: 10.1128/MCB.20.4.1254-1262.2000
Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9
Abstract
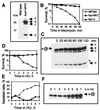
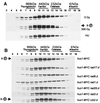
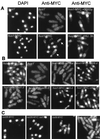
Hus1 is one of six checkpoint Rad proteins required for all Schizosaccharomyces pombe DNA integrity checkpoints. MYC-tagged Hus1 reveals four discrete forms. The main form, Hus1-B, participates in a protein complex with Rad9 and Rad1, consistent with reports that Rad1-Hus1 immunoprecipitation is dependent on the rad9(+) locus. A small proportion of Hus1-B is intrinsically phosphorylated in undamaged cells and more becomes phosphorylated after irradiation. Hus1-B phosphorylation is not increased in cells blocked in early S phase with hydroxyurea unless exposure is prolonged. The Rad1-Rad9-Hus1-B complex is readily detectable, but upon cofractionation of soluble extracts, the majority of each protein is not present in this complex. Indirect immunofluorescence demonstrates that Hus1 is nuclear and that this localization depends on Rad17. We show that Rad17 defines a distinct protein complex in soluble extracts that is separate from Rad1, Rad9, and Hus1. However, two-hybrid interaction, in vitro association and in vivo overexpression experiments suggest a transient interaction between Rad1 and Rad17.
Figures



References
-
- Bähler J, Wu J Q, Longtine M S, Shah N G, McKenzie A, Steever A B, Wach A, Philippsen P, Pringle J R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
-
- Boddy M N, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
