Fusions with histone H3 result in highly specific alteration of gene expression
- PMID: 10648797
- PMCID: PMC102566
- DOI: 10.1093/nar/28.4.1026
Fusions with histone H3 result in highly specific alteration of gene expression
Abstract
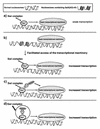
Hap1 is a yeast transcriptional activator which controls expression of genes such as CYC1 and CYC7. Our results show that Hap1 activity is dependent on a functional chromatin remodeling complex SWI/SNF. Using a modified two-hybrid screen with Hap1 as bait, we recovered expression vectors encoding the Gal4 activation domain fused to histone H3 [Gal4(AD)-H3]. Hap1 activity at CYC1 or CYC7 was increased by Gal4(AD)-H3 and the effect was dependent on the presence of the activation domain of Hap1 and a functional SWI complex. Importantly, overexpression of H3 alone had no effect on Hap1 activity. Analysis of Gal4(AD)-H3 revealed that the fusion is not incorporated into the nucleosome while a functional Gal4 activation domain is dispensable. Activity of many other transcriptional activators was unchanged or slightly affected in the presence of Gal4(AD)-H3. Thus, our results identify a new class of histone H3 variants that cause highly specific alteration of gene expression. Hap1 may interact directly with H3 favoring chromatin remodeling by the SWI/SNF complex.
Figures







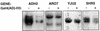
Similar articles
-
Mutations in target DNA elements of yeast HAP1 modulate its transcriptional activity without affecting DNA binding.Nucleic Acids Res. 1996 Apr 15;24(8):1453-9. doi: 10.1093/nar/24.8.1453. Nucleic Acids Res. 1996. PMID: 8628677 Free PMC article.
-
Mutations that alter transcriptional activation but not DNA binding in the zinc finger of yeast activator HAPI.Nature. 1989 Nov 9;342(6246):200-3. doi: 10.1038/342200a0. Nature. 1989. PMID: 2509943
-
Structure of HAP1-PC7 bound to DNA: implications for DNA recognition and allosteric effects of DNA-binding on transcriptional activation.Nucleic Acids Res. 2000 Oct 15;28(20):3853-63. doi: 10.1093/nar/28.20.3853. Nucleic Acids Res. 2000. PMID: 11024163 Free PMC article.
-
Structure of HAP1-18-DNA implicates direct allosteric effect of protein-DNA interactions on transcriptional activation.Nat Struct Biol. 1999 Jan;6(1):22-7. doi: 10.1038/4893. Nat Struct Biol. 1999. PMID: 9886287
-
Role of the HAP1 protein in repair of oxidative DNA damage and regulation of transcription factors.Br J Cancer Suppl. 1996 Jul;27:S145-50. Br J Cancer Suppl. 1996. PMID: 8763868 Free PMC article. Review. No abstract available.
Cited by
-
Nucleosome distortion as a possible mechanism of transcription activation domain function.Epigenetics Chromatin. 2016 Sep 20;9:40. doi: 10.1186/s13072-016-0092-2. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27679670 Free PMC article. Review.
-
The heme activator protein Hap1 represses transcription by a heme-independent mechanism in Saccharomyces cerevisiae.Genetics. 2005 Mar;169(3):1343-52. doi: 10.1534/genetics.104.037143. Epub 2005 Jan 16. Genetics. 2005. PMID: 15654089 Free PMC article.
-
A fungal family of transcriptional regulators: the zinc cluster proteins.Microbiol Mol Biol Rev. 2006 Sep;70(3):583-604. doi: 10.1128/MMBR.00015-06. Microbiol Mol Biol Rev. 2006. PMID: 16959962 Free PMC article. Review.
References
-
- Wolffe A. (1998) Chromatin, Structure and Function. Academic Press, London, UK.
-
- Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Nature, 389, 251–260. - PubMed
-
- Clark-Adams C.D., Norris,D., Osley,M.A., Fassler,J.S. and Winston,F. (1988) Genes Dev., 2, 150–159. - PubMed
-
- Han M. and Grunstein,M. (1988) Cell, 55, 1137–1145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

