Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning
- PMID: 10652276
- PMCID: PMC316342
Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning
Abstract
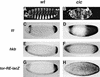
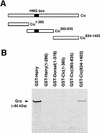
Differentiation of the embryonic termini in Drosophila depends on signaling by the Tor RTK, which induces terminal gene expression by inactivating at the embryonic poles a uniformly distributed repressor activity that involves the Gro corepressor. Here, we identify a new gene, cic, that acts as a repressor of terminal genes regulated by the Tor pathway. cic also mediates repression along the dorsoventral axis, a process that requires the Dorsal morphogen and Gro, and which is also inhibited by Tor signaling at the termini. cic encodes an HMG-box transcription factor that interacts with Gro in vitro. We present evidence that Tor signaling regulates terminal patterning by inactivating Cic at the embryo poles. cic has been evolutionarily conserved, suggesting that Cic-like proteins may act as repressors regulated by RTK signaling in other organisms.
Figures




References
-
- Brunner D, Oellers N, Szabad J, Biggs WH, Zipursky SL, Hafen E. A gain of function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signalling pathways. Cell. 1994;76:875–888. - PubMed
-
- Casanova J. Interaction between torso and dorsal, two elements of different transduction pathways in the Drosophila embryo. Mech Dev. 1991;36:41–45. - PubMed
-
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. - PubMed
-
- Chou TB, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–1369. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
