Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes
- PMID: 10653705
- PMCID: PMC91850
- DOI: 10.1128/AEM.66.2.467-475.2000
Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes
Abstract
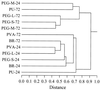
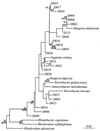
The nearly universal colonization of surfaces in marine waters by bacteria and the formation of biofilms and biofouling communities have important implications for ecological function and industrial processes. However, the dynamics of surface attachment and colonization in situ, particularly during the early stages of biofilm establishment, are not well understood. Experimental surfaces that differed in their degrees of hydrophilicity or hydrophobicity were incubated in a salt marsh estuary tidal creek for 24 or 72 h. The organisms colonizing these surfaces were examined by using a cultivation-independent approach, amplified ribosomal DNA restriction analysis. The goals of this study were to assess the diversity of bacterial colonists involved in early succession on a variety of surfaces and to determine the phylogenetic affiliations of the most common early colonists. Substantial differences in the representation of different cloned ribosomal DNA sequences were found when the 24- and 72-h incubations were compared, indicating that some new organisms were recruited and some other organisms were lost. Phylogenetic analyses of the most common sequences recovered showed that the colonists were related to organisms known to inhabit surfaces or particles in marine systems. A total of 22 of the 26 clones sequenced were affiliated with the Roseobacter subgroup of the alpha subdivision of the division Proteobacteria (alpha-Proteobacteria), and most of these clones were recovered at a high frequency from all surfaces after 24 or 72 h of incubation. Two clones were affiliated with the Alteromonas group of the gamma-Proteobacteria and appeared to be involved only in the very early stages of colonization (within the first 24 h). A comparison of the colonization patterns on the test surfaces indicated that the early bacterial community succession rate and/or direction may be influenced by surface physicochemical properties. However, organisms belonging to the Roseobacter subgroup are ubiquitous and rapid colonizers of surfaces in coastal environments.
Figures
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987.
-
- Baier R E, Shafrin E G, Zisman W A. Adhesion. Mechanisms that assist or impede it. Science. 1968;162:1360–1368. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

