Dynamics of bacterial community composition and activity during a mesocosm diatom bloom
- PMID: 10653721
- PMCID: PMC91866
- DOI: 10.1128/AEM.66.2.578-587.2000
Dynamics of bacterial community composition and activity during a mesocosm diatom bloom
Erratum in
- Appl Environ Microbiol 2000 May;66(5):2282
Abstract
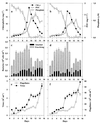
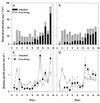
Bacterial community composition, enzymatic activities, and carbon dynamics were examined during diatom blooms in four 200-liter laboratory seawater mesocosms. The objective was to determine whether the dramatic shifts in growth rates and ectoenzyme activities, which are commonly observed during the course of phytoplankton blooms and their subsequent demise, could result from shifts in bacterial community composition. Nutrient enrichment of metazoan-free seawater resulted in diatom blooms dominated by a Thalassiosira sp., which peaked 9 days after enrichment ( approximately 24 microg of chlorophyll a liter(-1)). At this time bacterial abundance abruptly decreased from 2.8 x 10(6) to 0.75 x 10(6) ml(-1), and an analysis of bacterial community composition, by denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S rRNA gene fragments, revealed the disappearance of three dominant phylotypes. Increased viral and flagellate abundances suggested that both lysis and grazing could have played a role in the observed phylotype-specific mortality. Subsequently, new phylotypes appeared and bacterial production, abundance, and enzyme activities shifted from being predominantly associated with the <1.0-microm size fraction towards the >1.0-microm size fraction, indicating a pronounced microbial colonization of particles. Sequencing of DGGE bands suggested that the observed rapid and extensive colonization of particulate matter was mainly by specialized alpha-Proteobacteria- and Cytophagales-related phylotypes. These particle-associated bacteria had high growth rates as well as high cell-specific aminopeptidase, beta-glucosidase, and lipase activities. Rate measurements as well as bacterial population dynamics were almost identical among the mesocosms indicating that the observed bacterial community dynamics were systematic and repeatable responses to the manipulated conditions.
Figures



References
-
- Alldredge A L, Passow U, Logan B E. The abundance of a class of large, transparent organic particles in the ocean. Deep-Sea Res. 1993;40:1131–1140.
-
- Azam F. Microbial control of oceanic carbon flux: the plot thickens. Science. 1998;280:694–696.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

