Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution
- PMID: 10654930
- PMCID: PMC305568
- DOI: 10.1093/emboj/19.3.317
Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution
Abstract
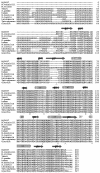
Fibrillarin is a phylogenetically conserved protein essential for efficient processing of pre-rRNA through its association with a class of small nucleolar RNAs during ribosomal biogenesis. The protein is the antigen for the autoimmune disease scleroderma. Here we report the crystal structure of the fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution. The structure consists of two domains, with a novel fold in the N-terminal region and a methyltransferase-like domain in the C-terminal region. Mapping temperature-sensitive mutations found in yeast fibrillarin Nop1 to the Methanococcus homologue structure reveals that many of the mutations cluster in the core of the methyltransferase-like domain.
Figures



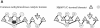
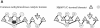
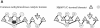

References
-
- Brünger A.T. (1992a) X-PLOR Version 3.1. A System for Crystallography and NMR. Yale University Press, New Haven, CT.
-
- Brünger A.T. (1992b) The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 335, 472–475. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

