Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice
- PMID: 10655499
- PMCID: PMC15551
- DOI: 10.1073/pnas.97.3.1148
Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice
Abstract
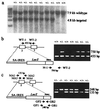
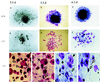
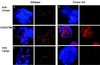
Centromere protein A (Cenpa for mouse, CENP-A for other species) is a histone H3-like protein that is thought to be involved in the nucleosomal packaging of centromeric DNA. Using gene targeting, we have disrupted the mouse Cenpa gene and demonstrated that the gene is essential. Heterozygous mice are healthy and fertile whereas null mutants fail to survive beyond 6.5 days postconception. Affected embryos show severe mitotic problems, including micronuclei and macronuclei formation, nuclear bridging and blebbing, and chromatin fragmentation and hypercondensation. Immunofluorescence analysis of interphase cells at day 5.5 reveals complete Cenpa depletion, diffuse Cenpb foci, absence of discrete Cenpc signal on centromeres, and dispersion of Cenpb and Cenpc throughout the nucleus. These results suggest that Cenpa is essential for kinetochore targeting of Cenpc and plays an early role in organizing centromeric chromatin at interphase. The evidence is consistent with the proposal of a critical epigenetic function for CENP-A in marking a chromosomal region for centromere formation.
Figures

References
-
- Choo K H A. The Centromere. Oxford: Oxford Univ. Press; 1997.
-
- Rattner J B. BioEssays. 1991;13:51–56. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

