The rice tungro bacilliform virus gene II product interacts with the coat protein domain of the viral gene III polyprotein
- PMID: 10666237
- PMCID: PMC111688
- DOI: 10.1128/jvi.74.5.2073-2083.2000
The rice tungro bacilliform virus gene II product interacts with the coat protein domain of the viral gene III polyprotein
Abstract
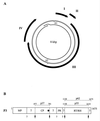
Rice tungro bacilliform virus (RTBV) is a plant pararetrovirus whose DNA genome contains four genes encoding three proteins and a large polyprotein. The function of most of the viral proteins is still unknown. To investigate the role of the gene II product (P2), we searched for interactions between this protein and other RTBV proteins. P2 was shown to interact with the coat protein (CP) domain of the viral gene III polyprotein (P3) both in the yeast two-hybrid system and in vitro. Domains involved in the P2-CP association have been identified and mapped on both proteins. To determine the importance of this interaction for viral multiplication, the infectivity of RTBV gene II mutants was investigated by agroinoculation of rice plants. The results showed that virus viability correlates with the ability of P2 to interact with the CP domain of P3. This study suggests that P2 could participate in RTBV capsid assembly.
Figures




Similar articles
-
Characterization of the genome of rice tungro bacilliform virus: comparison with Commelina yellow mottle virus and caulimoviruses.Virology. 1991 Nov;185(1):354-64. doi: 10.1016/0042-6822(91)90783-8. Virology. 1991. PMID: 1926781
-
Characterization of the protease domain of Rice tungro bacilliform virus responsible for the processing of the capsid protein from the polyprotein.Virol J. 2005 Apr 14;2:33. doi: 10.1186/1743-422X-2-33. Virol J. 2005. PMID: 15831103 Free PMC article.
-
Rice tungro bacilliform virus open reading frame 3 encodes a single 37-kDa coat protein.Virology. 1999 Jan 20;253(2):319-26. doi: 10.1006/viro.1998.9519. Virology. 1999. PMID: 9918890
-
Rice tungro bacilliform virus open reading frames II and III are translated from polycistronic pregenomic RNA by leaky scanning.J Virol. 1997 Oct;71(10):7984-9. doi: 10.1128/JVI.71.10.7984-7989.1997. J Virol. 1997. PMID: 9311892 Free PMC article.
-
A short basic domain supports a nucleic acid-binding activity in the rice tungro bacilliform virus open reading frame 2 product.Virology. 1997 Dec 22;239(2):352-9. doi: 10.1006/viro.1997.8859. Virology. 1997. PMID: 9434726
Cited by
-
Interaction between the open reading frame III product and the coat protein is required for transmission of cauliflower mosaic virus by aphids.J Virol. 2001 Jan;75(1):100-6. doi: 10.1128/JVI.75.1.100-106.2001. J Virol. 2001. PMID: 11119578 Free PMC article.
-
The product of ORF III in cauliflower mosaic virus interacts with the viral coat protein through its C-terminal proline rich domain.Virus Genes. 2001 Mar;22(2):159-65. doi: 10.1023/a:1008121228637. Virus Genes. 2001. PMID: 11324752
-
The molecular diversity and evolution of Rice tungro bacilliform virus from Indian perspective.Virus Genes. 2012 Aug;45(1):126-38. doi: 10.1007/s11262-012-0751-8. Epub 2012 Apr 29. Virus Genes. 2012. PMID: 22544477
-
Virus-induced gene silencing in rice using a vector derived from a DNA virus.Planta. 2010 Nov;232(6):1531-40. doi: 10.1007/s00425-010-1273-z. Epub 2010 Sep 25. Planta. 2010. PMID: 20872012
-
A coiled-coil interaction mediates cauliflower mosaic virus cell-to-cell movement.Proc Natl Acad Sci U S A. 2005 Apr 26;102(17):6219-24. doi: 10.1073/pnas.0407731102. Epub 2005 Apr 18. Proc Natl Acad Sci U S A. 2005. PMID: 15837934 Free PMC article.
References
-
- Bao Y, Hull R. Characterization of the discontinuities in rice tungro bacilliform virus DNA. J Gen Virol. 1992;73:1297–1301. - PubMed
-
- Cheng C P, Lockhart B E, Olszewski N E. The ORF I and II proteins of Commelina yellow mottle virus are virion-associated. Virology. 1996;223:263–271. - PubMed
-
- Dahal G, Hibino H, Saxena R C. Association of leafhopper feeding behavior with transmission of rice tungro to susceptible and resistant rice cultivars. Phytopathology. 1990;80:371–377.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

