Induction of the cellular E2F-1 promoter by the adenovirus E4-6/7 protein
- PMID: 10666238
- PMCID: PMC111689
- DOI: 10.1128/jvi.74.5.2084-2093.2000
Induction of the cellular E2F-1 promoter by the adenovirus E4-6/7 protein
Abstract
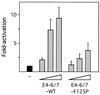
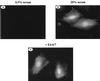
The adenovirus type 5 (Ad5) E4-6/7 protein interacts directly with different members of the E2F family and mediates the cooperative and stable binding of E2F to a unique pair of binding sites in the Ad5 E2a promoter region. This induction of E2F DNA binding activity strongly correlates with increased E2a transcription when analyzed using virus infection and transient expression assays. Here we show that while different adenovirus isolates express an E4-6/7 protein that is capable of induction of E2F dimerization and stable DNA binding to the Ad5 E2a promoter region, not all of these viruses carry the inverted E2F binding site targets in their E2a promoter regions. The Ad12 and Ad40 E2a promoter regions bind E2F via a single binding site. However, these promoters bind adenovirus-induced (dimerized) E2F very weakly. The Ad3 E2a promoter region binds E2F very poorly, even via a single binding site. A possible explanation of these results is that the Ad E4-6/7 protein evolved to induce cellular gene expression. Consistent with this notion, we show that infection with different adenovirus isolates induces the binding of E2F to an inverted configuration of binding sites present in the cellular E2F-1 promoter. Transient expression of the E4-6/7 protein alone in uninfected cells is sufficient to induce transactivation of the E2F-1 promoter linked to chloramphenicol acetyltransferase or green fluorescent protein reporter genes. Further, expression of the E4-6/7 protein in the context of adenovirus infection induces E2F-1 protein accumulation. Thus, the induction of E2F binding to the E2F-1 promoter by the E4-6/7 protein observed in vitro correlates with transactivation of E2F-1 promoter activity in vivo. These results suggest that adenovirus has evolved two distinct mechanisms to induce the expression of the E2F-1 gene. The E1A proteins displace repressors of E2F activity (the Rb family members) and thus relieve E2F-1 promoter repression; the E4-6/7 protein complements this function by stably recruiting active E2F to the E2F-1 promoter to transactivate expression.
Figures
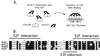




Similar articles
-
The adenovirus E4-6/7 protein directs nuclear localization of E2F-4 via an arginine-rich motif.J Virol. 2005 Feb;79(4):2301-8. doi: 10.1128/JVI.79.4.2301-2308.2005. J Virol. 2005. PMID: 15681431 Free PMC article.
-
The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex.Mol Cell Biol. 1994 Feb;14(2):1333-46. doi: 10.1128/mcb.14.2.1333-1346.1994. Mol Cell Biol. 1994. PMID: 8289811 Free PMC article.
-
Mutually exclusive interaction of the adenovirus E4-6/7 protein and the retinoblastoma gene product with internal domains of E2F-1 and DP-1.J Virol. 1994 Nov;68(11):6848-62. doi: 10.1128/JVI.68.11.6848-6862.1994. J Virol. 1994. PMID: 7933066 Free PMC article.
-
E1A 12S and 13S of the transformation-defective adenovirus type 12 strain CS-1 inactivate proteins of the RB family, permitting transactivation of the E2F-dependent promoter.J Virol. 1997 Dec;71(12):9538-48. doi: 10.1128/JVI.71.12.9538-9548.1997. J Virol. 1997. PMID: 9371617 Free PMC article.
-
Introduction to the E2F family: protein structure and gene regulation.Curr Top Microbiol Immunol. 1996;208:1-30. doi: 10.1007/978-3-642-79910-5_1. Curr Top Microbiol Immunol. 1996. PMID: 8575210 Review.
Cited by
-
Adenovirus E2F1 overexpression sensitizes LNCaP and PC3 prostate tumor cells to radiation in vivo.Int J Radiat Oncol Biol Phys. 2011 Feb 1;79(2):549-58. doi: 10.1016/j.ijrobp.2010.08.013. Int J Radiat Oncol Biol Phys. 2011. PMID: 21195876 Free PMC article.
-
Requirement of Sur2 for efficient replication of mouse adenovirus type 1.J Virol. 2004 Dec;78(23):12888-900. doi: 10.1128/JVI.78.23.12888-12900.2004. J Virol. 2004. PMID: 15542641 Free PMC article.
-
The adenovirus E4-6/7 protein directs nuclear localization of E2F-4 via an arginine-rich motif.J Virol. 2005 Feb;79(4):2301-8. doi: 10.1128/JVI.79.4.2301-2308.2005. J Virol. 2005. PMID: 15681431 Free PMC article.
-
The E4-6/7 protein functionally compensates for the loss of E1A expression in adenovirus infection.J Virol. 2000 Jul;74(13):5819-24. doi: 10.1128/jvi.74.13.5819-5824.2000. J Virol. 2000. PMID: 10846061 Free PMC article.
-
Evidence for involvement of BH3-only proapoptotic members in adenovirus-induced apoptosis.J Virol. 2007 Oct;81(19):10486-95. doi: 10.1128/JVI.00808-07. Epub 2007 Jul 25. J Virol. 2007. PMID: 17652400 Free PMC article.
References
-
- Adnane J, Shao Z, Robbins P D. The retinoblastoma susceptibility gene product represses transcription when directly bound to the promoter. J Biol Chem. 1995;270:8837–8843. - PubMed
-
- Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

