A novel RNA-binding protein from Triturus carnifex identified by RNA-ligand screening with the newt hammerhead ribozyme
- PMID: 10666442
- PMCID: PMC102618
- DOI: 10.1093/nar/28.5.1045
A novel RNA-binding protein from Triturus carnifex identified by RNA-ligand screening with the newt hammerhead ribozyme
Abstract
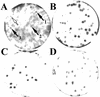
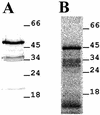
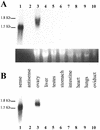
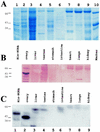
The newt hammerhead ribozyme is transcribed from Satellite 2 DNA, which consists of tandemly repeated units of 330 bp. However, different transcripts are synthesized in different tissues. In all somatic tissues and in testes, dimeric and multimeric RNA transcripts are generated which, to some extent, self-cleave into monomers at the hammerhead domain. In ovaries, primarily a distinct monomeric unit is formed by transcription, which retains an intact hammerhead self-cleavage site. The ovarian monomeric RNA associates to form a 12S complex with proteins that are poorly characterised so far. In this work we identified NORA, a protein that binds the ovarian form of the newt ribozyme. We show that the newt ribozyme binds to the Escherichia coli -expressed protein, as well as to a protein of identical size that is found exclusively in newt ovaries. Also NORA mRNA was detectable only in ovary, but in neither somatic tissues nor testes. The tissue-specific expression of NORA is analogous to the ovary-specific transcription of the newt ribozyme. Although NORA was identified by its ability to bind to the newt ribozyme in the presence of a vast excess of carrier RNA, it was able to interact with certain other RNA probes. This novel RNA-binding protein does not contain any motif characteristic for RNA-binding proteins or any other known protein domain, but it shares a striking similarity with a rat resiniferatoxin-binding protein.
Figures


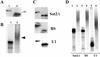
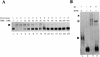
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

