A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis
- PMID: 10666445
- PMCID: PMC102592
- DOI: 10.1093/nar/28.5.1067
A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis
Abstract
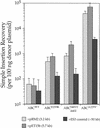
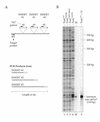
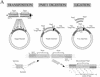
A robust Tn7-based in vitro transposition system is described that displays little target site selectivity, allowing the efficient recovery of many different transposon insertions in target DNAs ranging from small plasmids to cosmids to whole genomes. Two miniTn7 derivatives are described that are useful for the analysis of genes: one a derivative for making translational and transcriptional target gene fusions and the other a derivative that can generate 15 bp (5 amino acid) insertions in target DNAs (proteins).
Figures
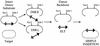






References
-
- Kleckner N., Bender,J. and Gottesman,S. (1991) Methods Enzymol., 204, 139–180. - PubMed
-
- Kleckner N., Roth,J. and Botstein,D. (1977) J. Mol. Biol., 116, 125–159. - PubMed
-
- Kaiser K., Sentry,J.W. and Finnegan,D.J. (1995) In Sherratt,D.J. (ed.), Eukaryotic Transposable Elements as Tools to Study Gene Structure and Function. IRL Press, Oxford, pp. 69–100.
-
- Berg C.M. and Berg,E. (1995) In Sherratt,D.J. (ed.), Transposable Elements as Tools for Molecular Analyses in Bacteria. IRL Press, Oxford, pp. 38–68.
-
- Waddell C.S. and Craig,N.L. (1988) Genes Dev., 2, 137–149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

