Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA
- PMID: 10669759
- PMCID: PMC85365
- DOI: 10.1128/MCB.20.5.1846-1854.2000
Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA
Abstract
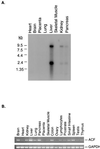
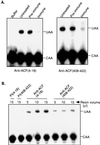
The C-to-U editing of apolipoprotein B (apo-B) mRNA is catalyzed by a multiprotein complex that recognizes an 11-nucleotide mooring sequence downstream of the editing site. The catalytic subunit of the editing enzyme, apobec-1, has cytidine deaminase activity but requires additional unidentified proteins to edit apo-B mRNA. We purified a 65-kDa protein that functionally complements apobec-1 and obtained peptide sequence information which was used in molecular cloning experiments. The apobec-1 complementation factor (ACF) cDNA encodes a novel 64.3-kDa protein that contains three nonidentical RNA recognition motifs. ACF and apobec-1 comprise the minimal protein requirements for apo-B mRNA editing in vitro. By UV cross-linking and immunoprecipitation, we show that ACF binds to apo-B mRNA in vitro and in vivo. Cross-linking of ACF is not competed by RNAs with mutations in the mooring sequence. Coimmunoprecipitation experiments identified an ACF-apobec-1 complex in transfected cells. Immunodepletion of ACF from rat liver extracts abolished editing activity. The immunoprecipitated complexes contained a functional holoenzyme. Our results support a model of the editing enzyme in which ACF binds to the mooring sequence in apo-B mRNA and docks apobec-1 to deaminate its target cytidine. The fact that ACF is widely expressed in human tissues that lack apobec-1 and apo-B mRNA suggests that ACF may be involved in other RNA editing or RNA processing events.
Figures





Similar articles
-
Identification of domains in apobec-1 complementation factor required for RNA binding and apolipoprotein-B mRNA editing.RNA. 2002 Jan;8(1):69-82. doi: 10.1017/s1355838202015649. RNA. 2002. PMID: 11871661 Free PMC article.
-
Identification of GRY-RBP as an apolipoprotein B RNA-binding protein that interacts with both apobec-1 and apobec-1 complementation factor to modulate C to U editing.J Biol Chem. 2001 Mar 30;276(13):10272-83. doi: 10.1074/jbc.M006435200. Epub 2000 Dec 27. J Biol Chem. 2001. PMID: 11134005
-
The editosome for cytidine to uridine mRNA editing has a native complexity of 27S: identification of intracellular domains containing active and inactive editing factors.J Cell Sci. 2002 Mar 1;115(Pt 5):1027-39. doi: 10.1242/jcs.115.5.1027. J Cell Sci. 2002. PMID: 11870221
-
RNA editing: cytidine to uridine conversion in apolipoprotein B mRNA.Biochim Biophys Acta. 2000 Nov 15;1494(1-2):1-13. doi: 10.1016/s0167-4781(00)00219-0. Biochim Biophys Acta. 2000. PMID: 11072063 Review.
-
Apobec-1 and apolipoprotein B mRNA editing.Biochim Biophys Acta. 1997 Mar 10;1345(1):11-26. doi: 10.1016/s0005-2760(96)00156-7. Biochim Biophys Acta. 1997. PMID: 9084497 Review.
Cited by
-
Biochemical and molecular characterization of two cytidine deaminases in the nematode Caenorhabditis elegans.Biochem J. 2002 Jul 1;365(Pt 1):99-107. doi: 10.1042/BJ20011814. Biochem J. 2002. PMID: 12071843 Free PMC article.
-
Transcriptome-wide organization of subcellular microenvironments revealed by ATLAS-Seq.Nucleic Acids Res. 2020 Jun 19;48(11):5859-5872. doi: 10.1093/nar/gkaa334. Nucleic Acids Res. 2020. PMID: 32421779 Free PMC article.
-
Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver.Genome Biol. 2014 Jun 19;15(6):R79. doi: 10.1186/gb-2014-15-6-r79. Genome Biol. 2014. PMID: 24946870 Free PMC article.
-
The Role of APOBECs in Viral Replication.Microorganisms. 2020 Nov 30;8(12):1899. doi: 10.3390/microorganisms8121899. Microorganisms. 2020. PMID: 33266042 Free PMC article. Review.
-
Transcriptional Regulation of Human Arylamine N-Acetyltransferase 2 Gene by Glucose and Insulin in Liver Cancer Cell Lines.Toxicol Sci. 2022 Nov 23;190(2):158-172. doi: 10.1093/toxsci/kfac103. Toxicol Sci. 2022. PMID: 36156098 Free PMC article.
References
-
- Anant S, Giannoni F, Antic D, DeMaria C T, Keene J D, Brewer G, Davidson N O. AU-rich RNA binding proteins Hel-N1 and AUF1 bind apolipoprotein B mRNA and inhibit posttranscriptional C to U editing. Nucleic Acids Symp Ser. 1997;36:115–118.
-
- Anant S, MacGinnitie A J, Davidson N O. apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, is a novel RNA-binding protein. J Biol Chem. 1995;270:14762–14767. - PubMed
-
- Bass B L. RNA editing and hypermutation by adenosine deamination. Trends Biochem Sci. 1997;22:157–162. . (Erratum, 22:278.) - PubMed
-
- Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
