Enhanced nitrogenase activity in strains of Rhodobacter capsulatus that overexpress the rnf genes
- PMID: 10671439
- PMCID: PMC94404
- DOI: 10.1128/JB.182.5.1208-1214.2000
Enhanced nitrogenase activity in strains of Rhodobacter capsulatus that overexpress the rnf genes
Abstract
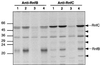
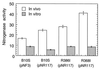
In the photosynthetic bacterium Rhodobacter capsulatus, a putative membrane-bound complex encoded by the rnfABCDGEH operon is thought to be dedicated to electron transport to nitrogenase. In this study, the whole rnf operon was cloned under the control of the nifH promoter in plasmid pNR117 and expressed in several rnf mutants. Complementation analysis demonstrated that transconjugants which integrated plasmid pNR117 directed effective biosynthesis of a functionally competent complex in R. capsulatus. Moreover, it was found that strains carrying pNR117 displayed nitrogenase activities 50 to 100% higher than the wild-type level. The results of radioactive labeling experiments indicated that the intracellular content of nitrogenase polypeptides was marginally altered in strains containing pNR117, whereas the levels of the RnfB and RnfC proteins present in the membrane were four- and twofold, respectively, higher than the wild-type level. Hence, the enhancement of in vivo nitrogenase activity was correlated with a commensurate overproduction of the Rnf polypeptides. In vitro nitrogenase assays performed in the presence of an artificial electron donor indicated that the catalytic activity of the enzyme was not increased in strains overproducing the Rnf polypeptides. It is proposed that the supply of reductants through the Rnf complex might be rate limiting for nitrogenase activity in vivo. Immunoprecipitation experiments performed on solubilized membrane proteins revealed that RnfB and RnfC are associated with each other and with additional polypeptides which may be components of the membrane-bound complex.
Figures




References
-
- Grabau C, Schatt E, Jouanneau Y, Vignais P M. A new [2Fe-2S] ferredoxin from Rhodobacter capsulatus. Coexpression with a 2[4Fe-4S] ferredoxin in Escherichia coli. J Biol Chem. 1991;266:3294–3299. - PubMed
-
- Jouanneau Y, Jeong H S, Hugo N, Meyer C, Willison J C. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus—characterization of two membrane-bound iron-sulfur proteins. Eur J Biochem. 1998;251:54–64. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

